| (1) HEART | (1) KIDNEY | (1) OTHER |
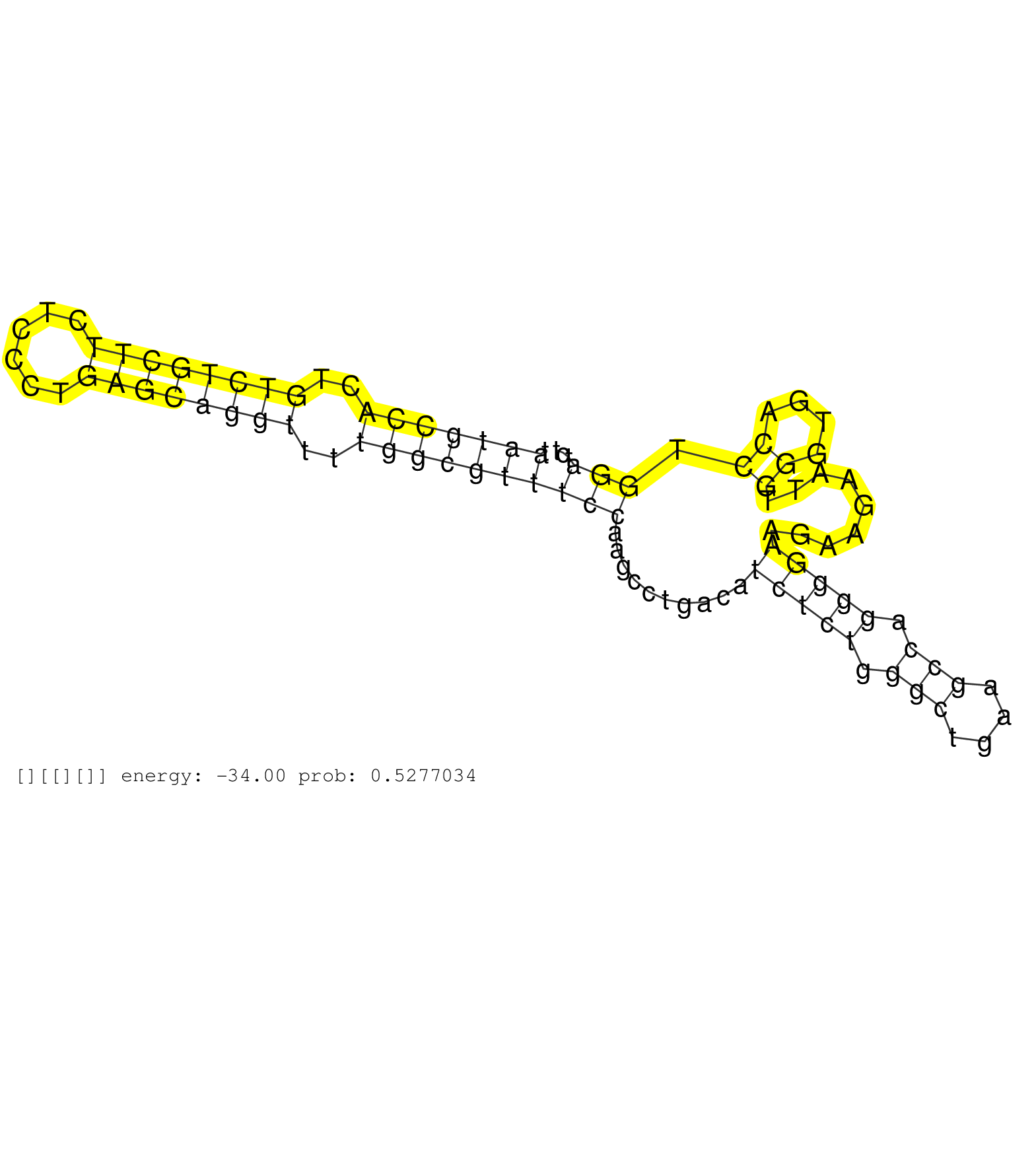
| AATGGGGATTAAGACTGTGAGCCCTATGTGGGACATGGACTGTCCAACCTGGTTAGCTTATATCTATCTGAGTGCTTAGTTCAGTGCCCAGCACGTAGTAAGTACTTAAATCCCATTTAAAAAAAAATGTGTCTCCCAGAAGTTGTCCTGTTTTATGGTGCAAGGGGTAGGGATTGGGCTCAGGGGAGCTCTCTGTGACCAGCTGATCCATAATGCCACTGTCTGCTTCTCCCTGAGCAGGTTTTGGCGTTTCCAAGCCTGACATCTCTGGGCTGAAGCCAGGGGAAGAAGAATTGGGTGACCTGGATCTTCAGGGCTCCAAGGATAAGGACATCCTGAGAGGCTCCTGT ...................................................................................................................................................................................................................(((((((..((((((((......))))))))..)))))))(((..........(((((.(((....))).))))).........((....)).)))........................................... ..................................................................................................................................................................................................................211................................................................................................310...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................CCACTGTCTGCTTCTCCCTGAGC................................................................................................................ | 23 | 1 | 4.00 | 4.00 | 4.00 | - | - |
| .......................................................................................................................................................................................................................CCACTGTCTGCTTCTCCTGAG.................................................................................................................. | 21 | 1.00 | 0.00 | - | - | 1.00 | |
| ............................................................................................................................................................................................................................................................................................GAAGAAGAATTGGGTGACCTGG............................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................................................................................................................................CCACTGTCTGCTTCTCCCTCAGC................................................................................................................ | 23 | 1.00 | 0.00 | - | 1.00 | - | |
| .....................................................................................................................................................................................................................................................................................................................................TAAGGACATCCTGAGAGGCT..... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ........................................................................................................................................................................................................................CACTGTCTGCTTCTCCCTGAGCAT.............................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .......................................................................................................................................................................................................................CCACTGTCTGCTTCTCCCTGAGCA............................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................................................................GACATCTCTGGGCTGA.......................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 |
| AATGGGGATTAAGACTGTGAGCCCTATGTGGGACATGGACTGTCCAACCTGGTTAGCTTATATCTATCTGAGTGCTTAGTTCAGTGCCCAGCACGTAGTAAGTACTTAAATCCCATTTAAAAAAAAATGTGTCTCCCAGAAGTTGTCCTGTTTTATGGTGCAAGGGGTAGGGATTGGGCTCAGGGGAGCTCTCTGTGACCAGCTGATCCATAATGCCACTGTCTGCTTCTCCCTGAGCAGGTTTTGGCGTTTCCAAGCCTGACATCTCTGGGCTGAAGCCAGGGGAAGAAGAATTGGGTGACCTGGATCTTCAGGGCTCCAAGGATAAGGACATCCTGAGAGGCTCCTGT ...................................................................................................................................................................................................................(((((((..((((((((......))))))))..)))))))(((..........(((((.(((....))).))))).........((....)).)))........................................... ..................................................................................................................................................................................................................211................................................................................................310...................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................ccGCTCAGGGAGAAGCC.............................................................................................................. | 17 | 1.00 | 0.00 | - | 1.00 | - | |
| ............................................................................................................................................................................................................................................................GAGATGTCAGGCTTGG.................................................................................. | 16 | 4 | 0.25 | 0.25 | - | - | 0.25 |
| ........................................................................TGCTGGGCACTGAACTAAGCA................................................................................................................................................................................................................................................................. | 21 | 7 | 0.14 | 0.14 | - | - | 0.14 |