| (1) HEART | (2) OTHER |
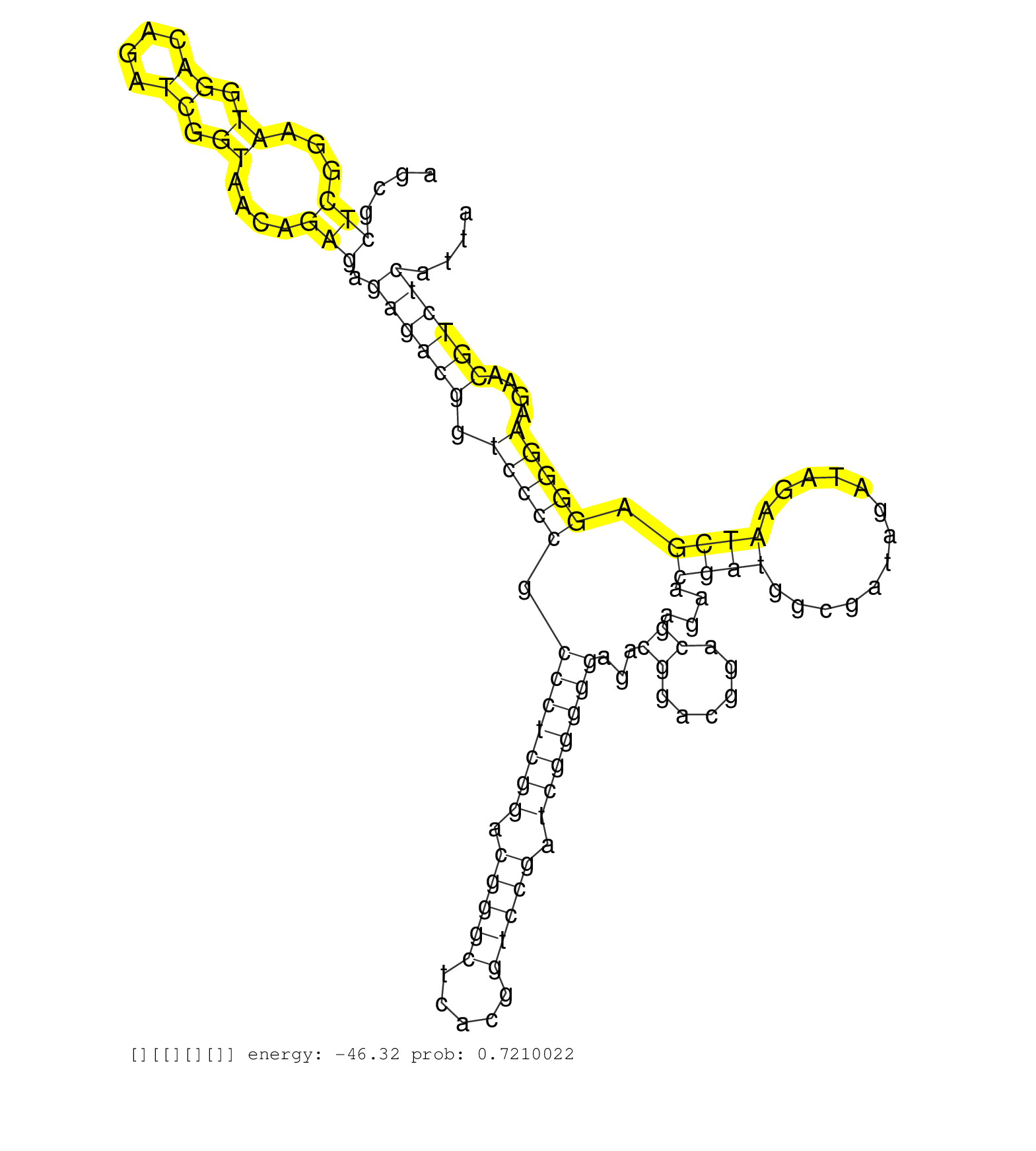
| GAAGCCAAGAAGAATCTCTGCGAAATCAACATCGACGACGACATGCCCAGGTAGCGCCCCGTCCCTCCTGCCGGGCGCAGCCGAGGGTCCGTCGGCAAGCCGGGGTTGCGGGGCTGCTGACGGTACCGTTCGTTCCGTGGTATTTATCGGGCGGAGCACTGTACTAAGCGCTCGGAATGGACAGATCGGTAACAGAGAGAGACGGTCCCCGCCCTCGGACGGGCTCACGGTCCGATCGGGGGAGACGGACGGACGAGAACGATGGCGATAGATAGAATCGAGGGGAAGAACGTCTCATTAAAACAATAGCAGAGAAATAGAATCAGGGTGATGTCCATCTCACCAACAAA ..........................................................................................................................................................................(((...((.((....)).))....))).((((((.(((((.(((((((.(((((.....))))).)))))))...((......))....((((.............)))).)))))....))))))...................................................... ......................................................................................................................................................................167..................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTAACGGT.......................................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | |
| ...................................................................................................................................................................CTAAGCGCTCGGAATGGACAGATA................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGCAT.............................................................................................................................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | |
| .......AGAAGAATCTCTGCGAAATCAACA............................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTAACGT........................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| ...............................................................................................................................................................................................................................................................................ATAGAATCGAGGGGAAGAAAAA......................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| ........................................................................................................................................................................................ATCGGTAACAGAGAGAGACGGAA............................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTACGAG........................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | |
| .....CAAGAAGAATCTCTGCGAAATCAACATCGAC.......................................................................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTAACAGGTGG....................................................................................................................................................... | 28 | 1.00 | 0.00 | 1.00 | - | - | |
| .........................................................................................................................................................................GCTCGGAATGGACAGATCCGCA............................................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | |
| ..........................................................................................................................................................................CTCGGAATGGACAGATCAAC................................................................................................................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | |
| ............................................................................................................................................................................................................................GGGCTCACGGTCCGATCGGGGACG.......................................................................................................... | 24 | 1.00 | 0.00 | - | 1.00 | - | |
| ..........................................................................................................................................................................CTCGGAATGGACAGATCA.................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | 1.00 | |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGA................................................................................................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTAACAGAGAGA...................................................................................................................................................... | 29 | 4 | 0.25 | 0.25 | 0.25 | - | - |
| ...........................................................................................................................................................................TCGGAATGGACAGATCGGTAACAGAGA........................................................................................................................................................ | 27 | 9 | 0.11 | 0.11 | 0.11 | - | - |
| GAAGCCAAGAAGAATCTCTGCGAAATCAACATCGACGACGACATGCCCAGGTAGCGCCCCGTCCCTCCTGCCGGGCGCAGCCGAGGGTCCGTCGGCAAGCCGGGGTTGCGGGGCTGCTGACGGTACCGTTCGTTCCGTGGTATTTATCGGGCGGAGCACTGTACTAAGCGCTCGGAATGGACAGATCGGTAACAGAGAGAGACGGTCCCCGCCCTCGGACGGGCTCACGGTCCGATCGGGGGAGACGGACGGACGAGAACGATGGCGATAGATAGAATCGAGGGGAAGAACGTCTCATTAAAACAATAGCAGAGAAATAGAATCAGGGTGATGTCCATCTCACCAACAAA ..........................................................................................................................................................................(((...((.((....)).))....))).((((((.(((((.(((((((.(((((.....))))).)))))))...((......))....((((.............)))).)))))....))))))...................................................... ......................................................................................................................................................................167..................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................................agatGCTATTGTTTTTAGA.................................... | 19 | 1.00 | 0.00 | - | 1.00 | - | |
| ............................................................................................................................................................................................................................................................agaCTATCGCCATCGTTCTAGA............................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - |