| (1) HEART | (1) KIDNEY | (2) OTHER |
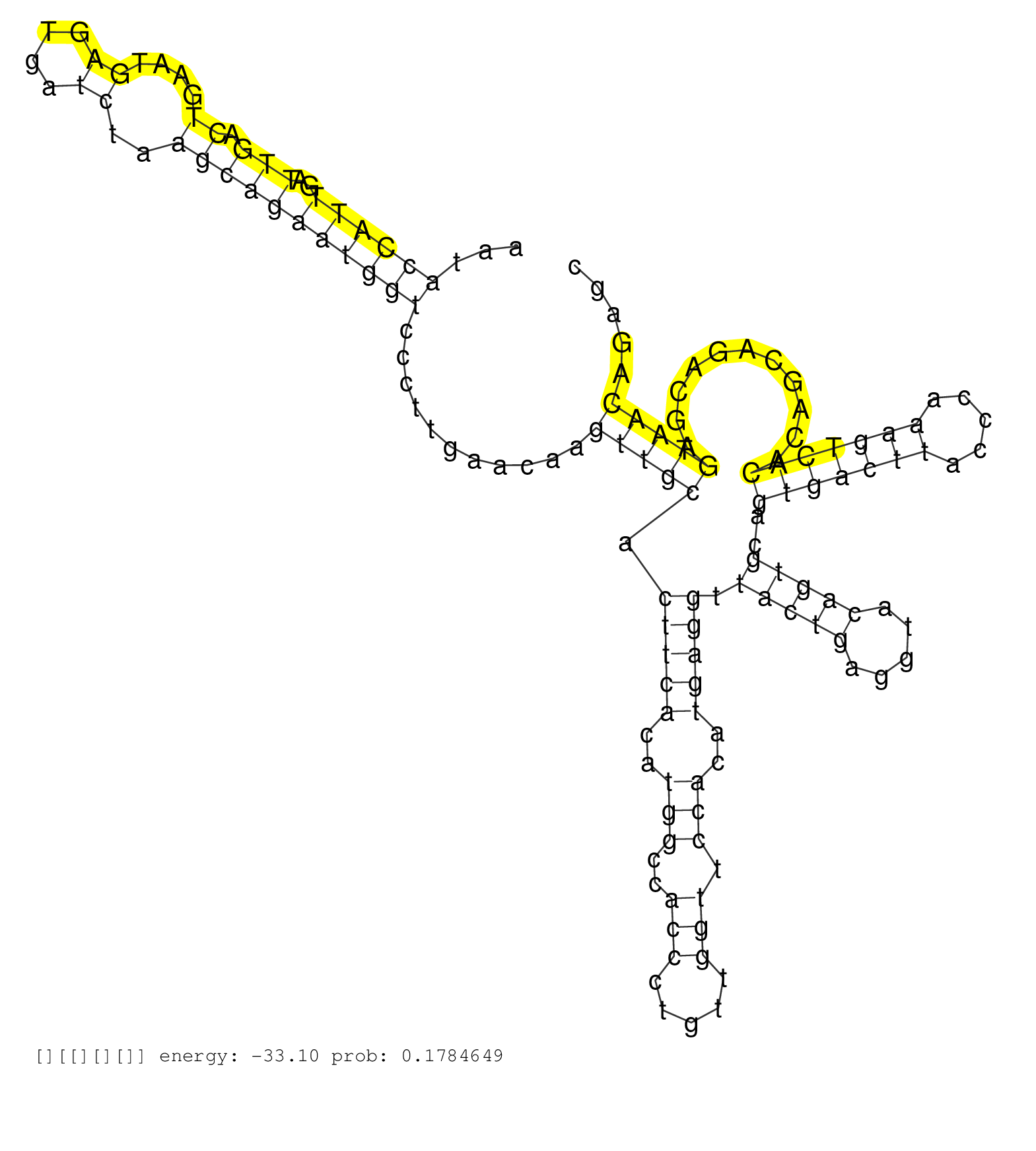
| CCTGCAGATCCGGGAGGTGTCCATCATGGAAAGCCAATAGGTGGAAGGAGGTATGGGAATCTTTTATCATTAAAGACACATCTGTGAATCTCTGAACTTCAAAAGAAAGAAGTAACCCATTAGTTATTACCTGCTGATGTCGCTCAGTAAATACCATTGATTGACTGAATGAGTGATCTAAGCAGAATGGTCCCTTGAACAAGTTGCACTTCACATGGCCACCCTGTTGGTTCCACATGAGGTTACTGAGGTACAGTGCAGTGACTTACCCAAAGTCACACAGCAGACGAGTAACAGAGCTGGGAATAGAATTCAAGTCCTTCTGACTCTCAGGCCTGTGTTCTCTCATT ........................................................................................................................................................((((((..(((.((....((....))..)))))))))))...........(((((.(((((..(((..(((.....))).)))..))))).(((((.....)))))..(((((((.....)))))))...........)))))....................................................... .....................................................................................................................................................150...................................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................CATTGATTGACTGAATGGGT................................................................................................................................................................................ | 20 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ..........................................................................................................................................................CATTGATTGACTGAATGGGAA............................................................................................................................................................................... | 21 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ..........................................................................................................................................................CATTGATTGACTGAATGGA................................................................................................................................................................................. | 19 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ..............................................................................................................................................................GATTGACTGAATGAGTGATCTAAGC....................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................TACCATTGATTGACTGAATGAGTGATCTAAGC....................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ........................................................................AAGACACATCTGTGAATCTCTGAACTT........................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ........................................................................................................................................ATGTCGCTCAGTAAATACC................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................................................................................................................................................CATTGATTGACTGAATGAGGAG.............................................................................................................................................................................. | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................................................................................................................................................................................................................................TCACACAGCAGACGAGTAAAAA..................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..........................................................................................................................................................CATTGATTGACTGAATGGAGG............................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..........................................................................................................................................................CATTGATTGACTGAATGGAG................................................................................................................................................................................ | 20 | 1.00 | 0.00 | 1.00 | - | - | - |
| CCTGCAGATCCGGGAGGTGTCCATCATGGAAAGCCAATAGGTGGAAGGAGGTATGGGAATCTTTTATCATTAAAGACACATCTGTGAATCTCTGAACTTCAAAAGAAAGAAGTAACCCATTAGTTATTACCTGCTGATGTCGCTCAGTAAATACCATTGATTGACTGAATGAGTGATCTAAGCAGAATGGTCCCTTGAACAAGTTGCACTTCACATGGCCACCCTGTTGGTTCCACATGAGGTTACTGAGGTACAGTGCAGTGACTTACCCAAAGTCACACAGCAGACGAGTAACAGAGCTGGGAATAGAATTCAAGTCCTTCTGACTCTCAGGCCTGTGTTCTCTCATT ........................................................................................................................................................((((((..(((.((....((....))..)))))))))))...........(((((.(((((..(((..(((.....))).)))..))))).(((((.....)))))..(((((((.....)))))))...........)))))....................................................... .....................................................................................................................................................150...................................................................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................gagaGTGGAACCAACAGAG.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ..................................................................................................................................................................................................................................................................................................................GGCCTGAGAGTCAGAAGGACTTGAATTCTA.............. | 30 | 4 | 0.25 | 0.25 | - | 0.25 | - | - |