| (1) HEART | (1) KIDNEY | (2) OTHER |
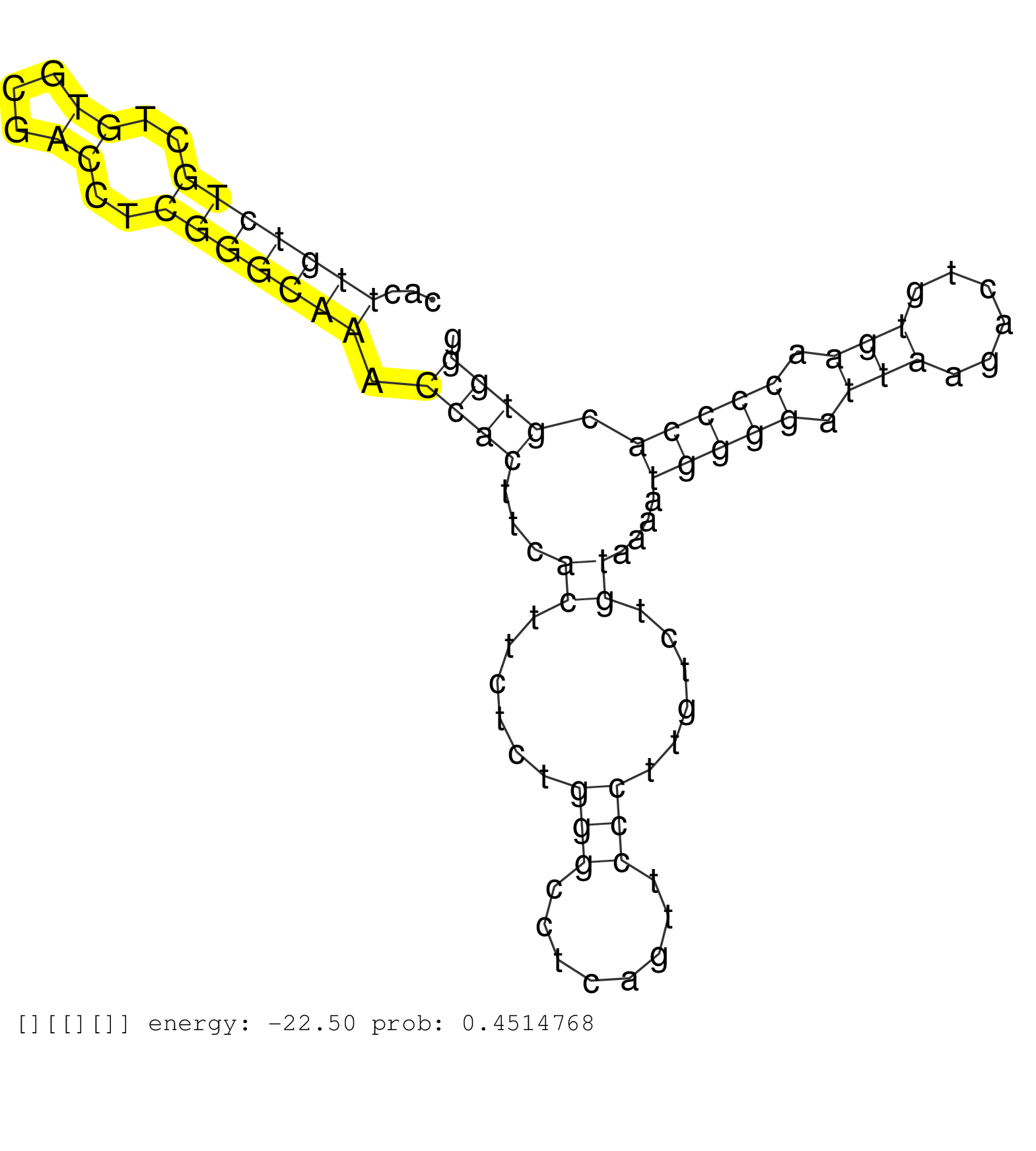
| CCAGTGTCTCGCCGGCTACCACCTCTCCCGTGACCGGAGCCTCTGCGAGGGTGAGCAGGGCCGGGTCGGGAGGTAGGGGCCGAGACAGACCCCCTCCCCCCCTCCGTTATTTCATCCTCTGGACTGGCCTAGGTCACAGTAGATACCACTGATTGACTGGTCAGAGCGACTAAAGAAGGGTGTTCTAATCCCGACTCCACCACTTGTCTGCTGTGCGACCTCGGGCAAACCACTTCACTTCTCTGGGCCTCAGTTCCCTTGTCTGTAAAATGGGGATTAAGACTGTGAACCCCACGTGGGACAACTCGCCACGAACGCTTGGATAAAGGGCTGTCTTCAATGTGATCCTG ...........................................................................................................................................................................................................(((((((..((...))..))))))).((((...((......(((........)))......))....(((((.(((......))).))))).))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCGAGA........................................................................................................................ | 22 | 4.00 | 0.00 | 4.00 | - | - | - | |
| ..............................................................................................................................................................................................................TCTGCTGTGCGACCTCGGGCGAGA........................................................................................................................ | 24 | 3.00 | 0.00 | 3.00 | - | - | - | |
| ............................CGTGACCGGAGCCTCTGC................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 |
| ..............................................................................................................................................................................................................TCTGCTGTGCGACCTCGGGCGAAA........................................................................................................................ | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCGAA......................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCGT.......................................................................................................................... | 20 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCGTGT........................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..............................................................................................................................................................................................................TCTGCTGTGCGACCTCGTA............................................................................................................................. | 19 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCATTA........................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ............................CGTGACCGGAGCCTCTGCGAGGA........................................................................................................................................................................................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | 1.00 | - | |
| .............................GTGACCGGAGCCTCTGCGA.............................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TGCTGTGCGACCTCGGGCGAAT........................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .......................................................................................................................................................ATTGACTGGTCAGAGC....................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | 0.25 | - | - |
| .......................................................CAGGGCCGGGTCGGG........................................................................................................................................................................................................................................................................................ | 15 | 8 | 0.12 | 0.12 | - | - | 0.12 | - |
| CCAGTGTCTCGCCGGCTACCACCTCTCCCGTGACCGGAGCCTCTGCGAGGGTGAGCAGGGCCGGGTCGGGAGGTAGGGGCCGAGACAGACCCCCTCCCCCCCTCCGTTATTTCATCCTCTGGACTGGCCTAGGTCACAGTAGATACCACTGATTGACTGGTCAGAGCGACTAAAGAAGGGTGTTCTAATCCCGACTCCACCACTTGTCTGCTGTGCGACCTCGGGCAAACCACTTCACTTCTCTGGGCCTCAGTTCCCTTGTCTGTAAAATGGGGATTAAGACTGTGAACCCCACGTGGGACAACTCGCCACGAACGCTTGGATAAAGGGCTGTCTTCAATGTGATCCTG ...........................................................................................................................................................................................................(((((((..((...))..))))))).((((...((......(((........)))......))....(((((.(((......))).))))).))))................................................... ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................tgcTGCCCGAGGTCGCACAGCAGACCGT........................................................................................................................ | 28 | 1.00 | 0.00 | 1.00 | - | - | - |