| (1) HEART | (1) KIDNEY |
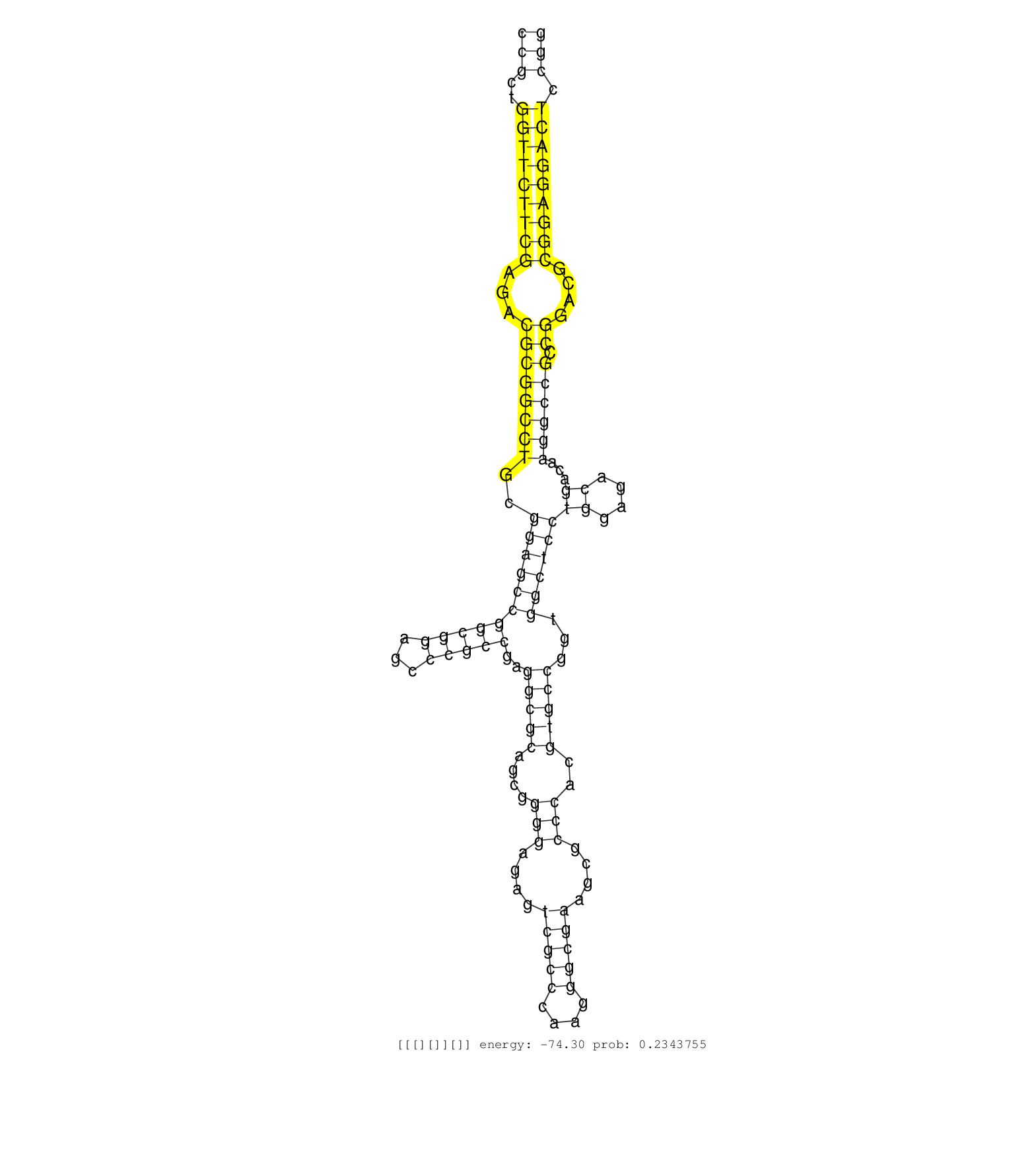
| GGAGGTCATCCGGGGCCTCACCCGCGAGGAGGCGGCGGCCGAGGACGGAAGGACGGCCCGCTGGTTCTTCGAGACGCGGCCTGCGGAGCCGGCGGAGCCCGCCGAGGCGCAGCGGGGAGAGTCGCCCAAGGGCGAAGCGCCCACGTGCCGGTGGCTCCTGGAGACGACAAGGCCGCCGGACGCGGAGGACTCCGGCGACCCCCGGGTCTCCGTGACCCCCGAGCCCTGCACGTGGATGCTGGAGGCGCAGCCCCCGGACGGGCAGGGGCCACCGCGCGAGCAACGTCTGCGGGCCGGCCAGGTGTCCGGCGTGGGCGGCGGGGTGGGCGGCACCGTCCGTCGCCTCTTCGAGACCGAGGCCCTGGGCGGCCAGGGCGGGCCCAGGAAGGCCGTTCGCACCTG .........................................................(((..(((((((((...((((((((..(((((((((((...)))))..(((((....(((....(((((....)))))....)))..)))))...))))))((....))...)))))).))....))))))))).)))............................................................................................................................................................................................................... .........................................................58.......................................................................................................................................195............................................................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ................................................................................................................................................................GAGACGACAAGGCCGCCGGACGCGGAGGACT................................................................................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | 2.00 | - |
| ..............................................................................................................................................................................GCCGGACGCGGAGGACT................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 |
| ....................................................................................................................................................................................................................................CACGTGGATGCTGGAG.............................................................................................................................................................. | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| ......................................................................................................................................................................ACAAGGCCGCCGGACGCGGAGGA..................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 |
| ...............................GCGGCGGCCGAGGACGGAAGGAC............................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - |
| ......................................................................................................................................................................ACAAGGCCGCCGGACGCGGA........................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 |
| .............................................................TGGTTCTTCGAGACGC..................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| ................................CGGCGGCCGAGGACGAGA................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | 1.00 | - | |
| .................................................................................................................................................................................GGACGCGGAGGACTCCG................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | 1.00 | - |
| ......................................................................................................................................................................................................................................CGTGGATGCTGGAGGC............................................................................................................................................................ | 16 | 1 | 1.00 | 1.00 | 1.00 | - |
| ........................................................................................................................................................................AAGGCCGCCGGACGCGGAGGACTCCGGC.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - |
| ......................................................................................................................................................................ACAAGGCCGCCGGACGCG.......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - |
| ..............................................................GGTTCTTCGAGACGCGGCCCG............................................................................................................................................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | |
| ............................................................................................................................................................................................................................................................................................GTCTGCGGGCCGGCCAGGTGTCCGGCGTGGGC...................................................................................... | 32 | 1 | 1.00 | 1.00 | 1.00 | - |
| ........................................................................................................................................................................................................................................................................................CAACGTCTGCGGGCCG.......................................................................................................... | 16 | 2 | 0.50 | 0.50 | 0.50 | - |
| GGAGGTCATCCGGGGCCTCACCCGCGAGGAGGCGGCGGCCGAGGACGGAAGGACGGCCCGCTGGTTCTTCGAGACGCGGCCTGCGGAGCCGGCGGAGCCCGCCGAGGCGCAGCGGGGAGAGTCGCCCAAGGGCGAAGCGCCCACGTGCCGGTGGCTCCTGGAGACGACAAGGCCGCCGGACGCGGAGGACTCCGGCGACCCCCGGGTCTCCGTGACCCCCGAGCCCTGCACGTGGATGCTGGAGGCGCAGCCCCCGGACGGGCAGGGGCCACCGCGCGAGCAACGTCTGCGGGCCGGCCAGGTGTCCGGCGTGGGCGGCGGGGTGGGCGGCACCGTCCGTCGCCTCTTCGAGACCGAGGCCCTGGGCGGCCAGGGCGGGCCCAGGAAGGCCGTTCGCACCTG .........................................................(((..(((((((((...((((((((..(((((((((((...)))))..(((((....(((....(((((....)))))....)))..)))))...))))))((....))...)))))).))....))))))))).)))............................................................................................................................................................................................................... .........................................................58.......................................................................................................................................195............................................................................................................................................................................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|