| (1) HEART | (1) KIDNEY | (2) OTHER |
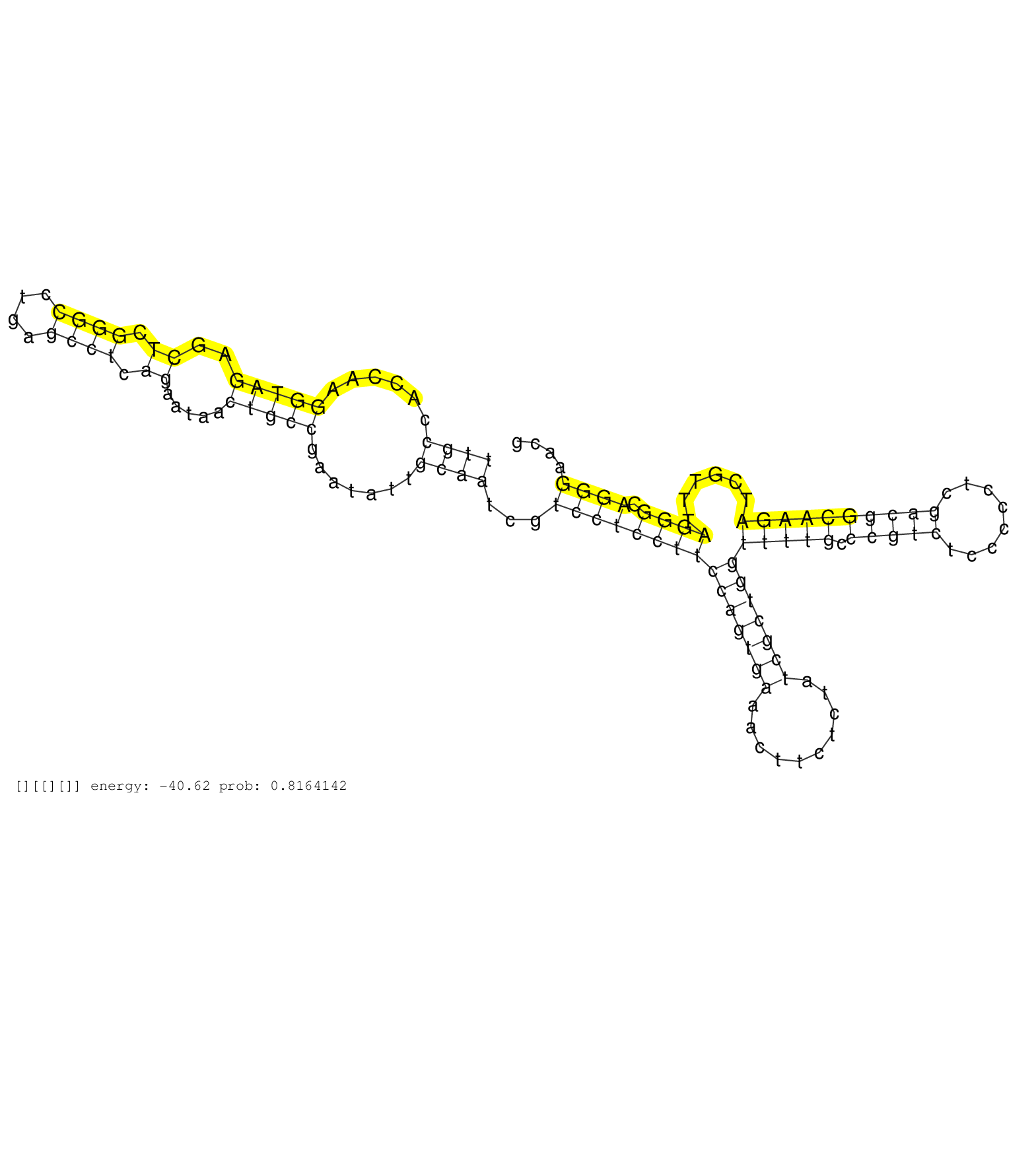
| TGCCCGGCGTGGGCACCGCCTACATCCAGAAGTACTCCAACGGGGGCAAGGTAAGGTTGGACACTCCCTCCTCCCCAACCTCCCATAGCCACCGAAGCCGCCCACAGACAAGCAGATAAGCAGCGTTGCCACCAAGGTAGAGCTCGGGCCTGAGCCTCAGAATAACTGCCGAATATTGCAATCGTCCTCCTTCCAGTGAAACTTCTCTATCGCTGGTTTTGCCCGTCTCCCCCTCGACGGCAAGATCGTTTAGGGCAGGGAACGTGTCTGCTAATTCTGTTGTACTCTCCCGAGCGCTCAGCACGGTGCTGGGCCCACACTAAGGGCTTCATTGATTGATTGAAACTGCC .............................................................................................................................((((......(((((..((.((((....)))).)).....))))).......))))...(((((((((((((((..........)))))))(((((.(((((........))))))))))......)))).)))).......................................................................................... .............................................................................................................................126.......................................................................................................................................264.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| ............................GAAGTACTCCAACGGGGGC............................................................................................................................................................................................................................................................................................................... | 19 | 1 | 3.00 | 3.00 | 3.00 | - | - | - |
| ..................................................................................................................................ACCAAGGTAGAGCTCGGGC......................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | 2.00 | - | - |
| ...........................AGAAGTACTCCAACGGGGGCAAGC........................................................................................................................................................................................................................................................................................................... | 24 | 2.00 | 0.00 | 2.00 | - | - | - | |
| ............................GAAGTACTCCAACGGGGGCAAGCA.......................................................................................................................................................................................................................................................................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ..................CCTACATCCAGAAGTACTCCAACGGGGGCAAGC........................................................................................................................................................................................................................................................................................................... | 33 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .............CACCGCCTACATCCAGAAGTACTCCAAC..................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .....................................................................................................................................AAGGTAGAGCTCGGGCC........................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...............................................................................................................................................................................................................................................GCAAGATCGTTTAGGGCAGGG.......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................CGCCTACATCCAGAAGTACT.......................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..............................................................................................................................................................AGAATAACTGCCGAATATTGCAATCGTCC................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................................................................TTCTGTTGTACTCTCCCGAGTATT.................................................... | 24 | 1.00 | 0.00 | - | - | - | 1.00 | |
| ............................................................................................................................................................................................................................................................................................................GCACGGTGCTGGGCC................................... | 15 | 5 | 0.20 | 0.20 | 0.20 | - | - | - |
| TGCCCGGCGTGGGCACCGCCTACATCCAGAAGTACTCCAACGGGGGCAAGGTAAGGTTGGACACTCCCTCCTCCCCAACCTCCCATAGCCACCGAAGCCGCCCACAGACAAGCAGATAAGCAGCGTTGCCACCAAGGTAGAGCTCGGGCCTGAGCCTCAGAATAACTGCCGAATATTGCAATCGTCCTCCTTCCAGTGAAACTTCTCTATCGCTGGTTTTGCCCGTCTCCCCCTCGACGGCAAGATCGTTTAGGGCAGGGAACGTGTCTGCTAATTCTGTTGTACTCTCCCGAGCGCTCAGCACGGTGCTGGGCCCACACTAAGGGCTTCATTGATTGATTGAAACTGCC .............................................................................................................................((((......(((((..((.((((....)))).)).....))))).......))))...(((((((((((((((..........)))))))(((((.(((((........))))))))))......)))).)))).......................................................................................... .............................................................................................................................126.......................................................................................................................................264.................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|