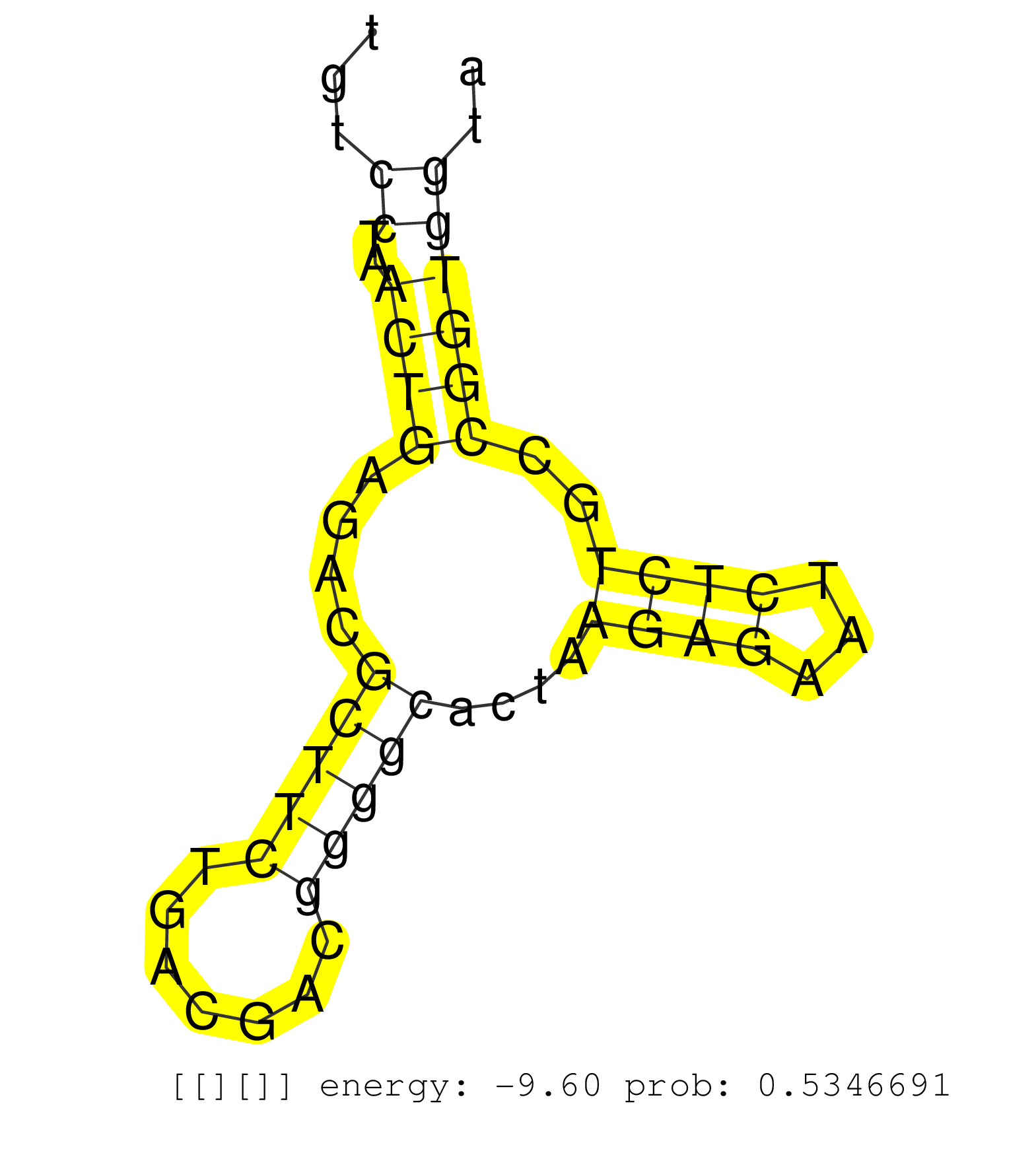
| (1) HEART | (1) KIDNEY | (2) OTHER |
| TTTTCTTTTGTGTGCGTGTGTTGTGGGGTGGTGGTTCTTTGGGTTTTTTTGTGCGTTTGTGCGTCTCCCCCGCACCCTCCCGGAAGACTCCCTCCTCCCCTTAAACGGACGCCAAATTGTCCCTCGAAAGTTGCGTGCGGCCGACCTTTTCACTTGTCCTAACTGAGACGCTTCTGACGACGGGGCACTAAGAGAATCTCTGCCGGTGGTAAAACCCAAATGCGTTTCTCCAGCTGTTCAGCGTGCGTAGAGAGGAAGGTGCTAACTGAGAGCCCGGGTCCCTGTTGCTTGGAGCCGCTGAGTCCTTGGAAGCCCACCGGTGGAGAGGTGGGCCGCAACGCCTCGGTCGT .............................................................................................................................................................((..((((....(((((.......)))))....((((...))))..))))))............................................................................................................................................. ..........................................................................................................................................................155.....................................................211......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TAACTGAGACGCTTCTGACGACGGGGCACTAAG.............................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ............................................................................................................................................................TCCTAACTGAGACGCTTCTGACGACGGGT..................................................................................................................................................................... | 29 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ...................................................................................................................................................TTTCACTTGTCCTAACTGAGACGCTTCTGA............................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................................................................................TGCTTGGAGCCGCTGAGTCCTTGGAAGCCC................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .............................................................................................................................................................................................AAGAGAATCTCTGCCGGT............................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................CAAATGCGTTTCTCCAGCTGTTCAGCGTG......................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ........TGTGTGCGTGTGTTGTGTGTT................................................................................................................................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | 1.00 | - | |
| ......................................................................................................................................................................................GGGCACTAAGAGAATCTCTGC................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................................................................................TGACGACGGGGCACTAAGAGAATCTCTGC................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ..........................................................................................................................................................TGTCCTAACTGAGACGCTTCTGACGACG........................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............................................................................................................................................................TAACTGAGACGCTTCTGACGACGGGGCAC.................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ....................................................................................................................................................TTCACTTGTCCTAACTGAGACGCTTCTGAC............................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............................................................................................................................................................TAACTGAGACGCTTCTGACGAC......................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 |
| ...........................................................................................................................................................GTCCTAACTGAGACGCTTCTGACGACG........................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| TTTTCTTTTGTGTGCGTGTGTTGTGGGGTGGTGGTTCTTTGGGTTTTTTTGTGCGTTTGTGCGTCTCCCCCGCACCCTCCCGGAAGACTCCCTCCTCCCCTTAAACGGACGCCAAATTGTCCCTCGAAAGTTGCGTGCGGCCGACCTTTTCACTTGTCCTAACTGAGACGCTTCTGACGACGGGGCACTAAGAGAATCTCTGCCGGTGGTAAAACCCAAATGCGTTTCTCCAGCTGTTCAGCGTGCGTAGAGAGGAAGGTGCTAACTGAGAGCCCGGGTCCCTGTTGCTTGGAGCCGCTGAGTCCTTGGAAGCCCACCGGTGGAGAGGTGGGCCGCAACGCCTCGGTCGT .............................................................................................................................................................((..((((....(((((.......)))))....((((...))))..))))))............................................................................................................................................. ..........................................................................................................................................................155.....................................................211......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TCTACGCACGCTGAACAGCTGGAGAAACGC................................................................................................... | 30 | 1 | 2.00 | 2.00 | 2.00 | - | - | - |
| ....................................................................................................................................CAGTTAGGACAAGTGAAAAGGTCGGCCGCACGC......................................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ......................................................................................................................................................TCAGAAGCGTCTCAGTTAGGACAAGTG............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .......................................................................................................................................................TCAGAAGCGTCTCAGTTAGGACAAGT............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |