| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
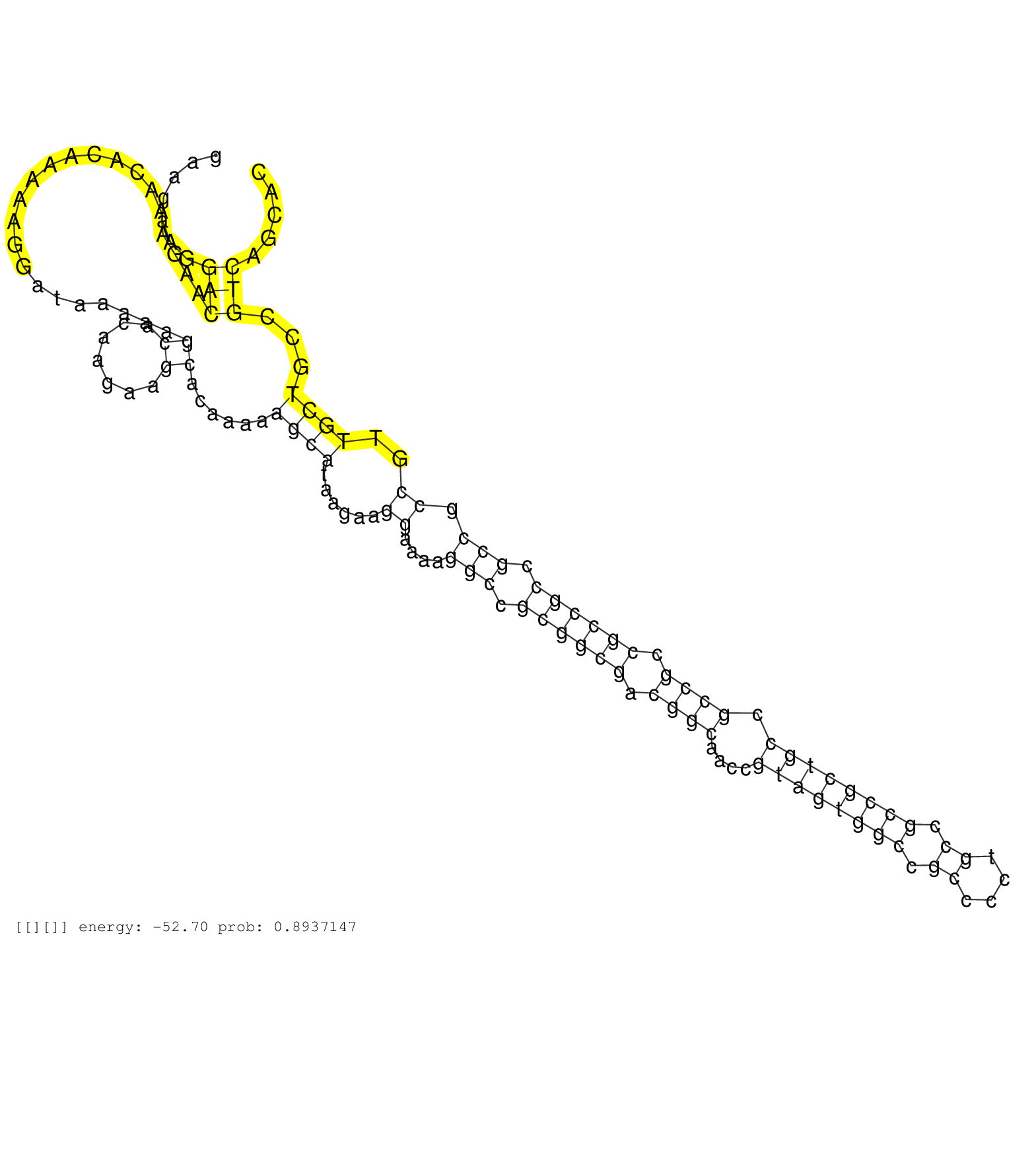
| GACGGGGCCTCCGTTTCCGTTCTTCCAGAATTCCGACCAAGAAGGCGGCGGGAAGAAGAAGAAGAAGAAGAAGGACAAGAAACACAAAAAGGATAAAAAGCACAAGAAGCACAAAAAGCATAAGAAGGAAAAGGCCGCGGCGACGGCAACCGTAGTGGCCGCCCCTGCCGCCGCTGCCGCCGCCGCCGCCGCCGCCGTTGCTGCCGTCAGCACGTCAGTCCAGGAAGAACAGGAGGCAGAAGCAGAGCCCAAAAAGGTGGGTG .........................................................................(((.......................((.......))......((((......((....(((.((((((.((((....((((((((.((....)).)))))))).)))).)))))).))).))..))))...)))....................................................... ..................................................................67................................................................................................................................................213................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................GTCAGCACGTCAGTCCAGGAAGAACAGGAGGCA......................... | 33 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - |
| ............................................................GAAGAAGAAGAAGGACAAGAAACACA................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................GTCAGTCCAGGAAGAACAGGAG............................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................GCACGTCAGTCCAGGAAGAACAGGAGGCA......................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................CAGGAAGAACAGGAGGCAGAAGCA................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................CACGTCAGTCCAGGAAGAACAGGAGGC.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| .....................................................................................................................................................................................................................GTCAGTCCAGGAAGAACAG............................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................................ACGTCAGTCCAGGAAGAACAGGAGGCAGAAGC.................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................AGCACGTCAGTCCAGGAAGAACAGG.............................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................CAGGAAGAACAGGAGGCAGAAGC.................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ...............................................GCGGGAAGAAGAAGAAGAAGAAGAAGGACA.......................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................AAGGACAAGAAACACAAAAAGGATAAAAAGC.................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................CGTCAGCACGTCAGTCCAGGAAGAACAGG.............................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................GCACGTCAGTCCAGGAAGAACAGGAGGCAG........................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................GCACGTCAGTCCAGGAAGAACAGGAGGC.......................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .......................................................................AGGACAAGAAACACAAAAAGG........................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................GTCAGCACGTCAGTCCAGGAAGAACA................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ......................................................................AAGGACAAGAAACACAAAAAGGATAAAAA.................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..................................................................................CACAAAAAGGATAAAAAGC.................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| .......................................................................AGGACAAGAAACACAAAAAGCAT......................................................................................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ...............................................................................................................CAAAAAGCATAAGAAGGAAAAGGCCGC............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................GTCAGTCCAGGAAGAACAGGAGGCAGAAGCAGA................. | 33 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ......................................................................................AAAAGGATAAAAAGCACAAGAAGCACAAAAA.................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................GTTGCTGCCGTCAGCACG................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .........................................................GAAGAAGAAGAAGAAGGACAAGAAA..................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .....................................................................GAAGGACAAGAAACACAAAAAGG........................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................................................GCAGAGCCCAAAAAGGAGAC.. | 20 | 1.00 | 0.00 | 1.00 | - | - | - | - | |
| ..............................................................................................................................................................................................................................GGAAGAACAGGAGGCAGAAGCA................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................CAGGAAGAACAGGAGGC.......................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - |
| .....................................................................GAAGGACAAGAAACAC.................................................................................................................................................................................. | 16 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - |
| GACGGGGCCTCCGTTTCCGTTCTTCCAGAATTCCGACCAAGAAGGCGGCGGGAAGAAGAAGAAGAAGAAGAAGGACAAGAAACACAAAAAGGATAAAAAGCACAAGAAGCACAAAAAGCATAAGAAGGAAAAGGCCGCGGCGACGGCAACCGTAGTGGCCGCCCCTGCCGCCGCTGCCGCCGCCGCCGCCGCCGCCGTTGCTGCCGTCAGCACGTCAGTCCAGGAAGAACAGGAGGCAGAAGCAGAGCCCAAAAAGGTGGGTG .........................................................................(((.......................((.......))......((((......((....(((.((((((.((((....((((((((.((....)).)))))))).)))).)))))).))).))..))))...)))....................................................... ..................................................................67................................................................................................................................................213................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|