| (1) HEART | (1) KIDNEY | (1) OTHER |
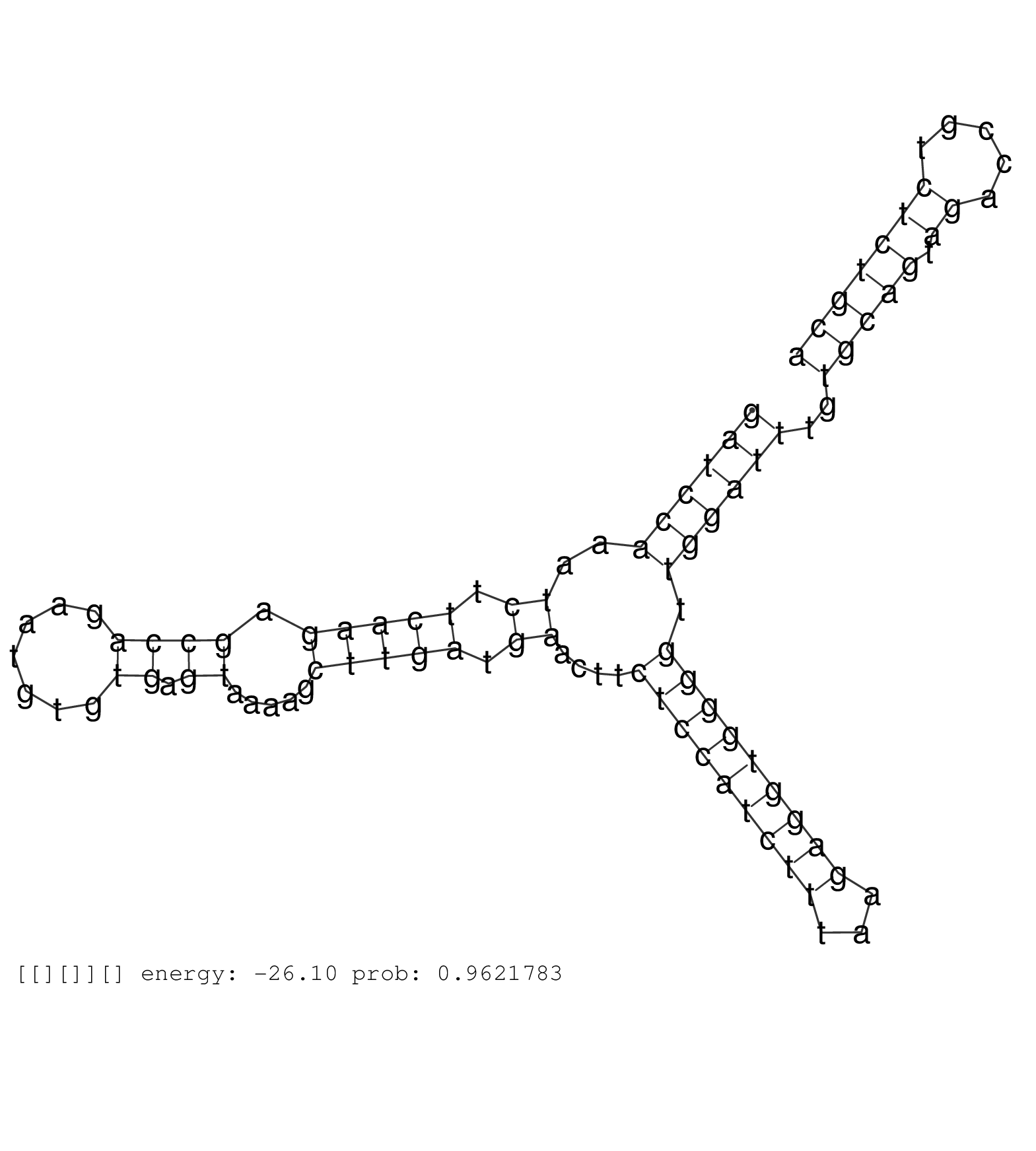
| AAATATGGCATAGGGAACAAATGAAGAGTGAATCCCGAGAGCAAAGAGAGGTGGGTTGGGAAGAAGTCATTGTTTTTTGCTTAGAGAAACTGGAAGCAGTTTCTTAAAGGTTGTGGTTTTTTGGTTTTTTTTAACAGGCAGAAGACAATTTGCGTAGAGAAAAAAACCTGGAAGAAGCAAAGAAGATTACCATTAAAAAAGATCCAAATCTTCAAGAGCCAGAATGTGTGAGTAAAAGCTTGATGAACTTCTCCATCTTTAAGAGGTGGGGTTGGATTTGTGCAGTAGACCGTCTCTGCAAAACAAAAACACCTCATCTCAGCTATTCTTCAGGCAAGCCCAAATAGTTC ........................................................................................................................................................................................................((((((..((.(((((.((((.......)).)).....))))).))....(((((((((...))))))))).))))))..(((((.((.....))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................ATTTGCGTAGAGAAAAAAACCTGGAAGAAGCA........................................................................................................................................................................... | 32 | 1 | 4.00 | 4.00 | 4.00 | - | - |
| .......................................................................................................................................................GCGTAGAGAAAAAAACCTGGAAGAAGC............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..............................................................................................................................................................GAAAAAAACCTGGAAGAAGCAAAGAAGA.................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .......................................................................................................................................................GCGTAGAGAAAAAAACCTGGAAGAAGCA........................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ..........TAGGGAACAAATGAAGAG.................................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................GTAGAGAAAAAAACCTGGAAG................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ......................................................................................................................................................TGCGTAGAGAAAAAAACCTGGAAGAAGCAA.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ......................................................................................................................................................TGCGTAGAGAAAAAAACCTGGAAGAAGCA........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| .......................................................................................................................................................GCGTAGAGAAAAAAACCTGGAAGAAGCAAA......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................GTAGAGAAAAAAACCTGGAAGAAGCAAAGAAG..................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...........................................................................................................................................................AGAGAAAAAAACCTGGAAGAAGCAAAGAAGAT................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...TATGGCATAGGGAACA........................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .........................................................................................................................................................GTAGAGAAAAAAACCTGGAAGAAGCAA.......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................................................ACAATTTGCGTAGAG............................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................AACCTGGAAGAAGCAAAGAAGATT.................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| .................................................................................................................................................................................................................................................................................GGATTTGTGCAGTAG.............................................................. | 15 | 6 | 0.17 | 0.17 | 0.17 | - | - |
| AAATATGGCATAGGGAACAAATGAAGAGTGAATCCCGAGAGCAAAGAGAGGTGGGTTGGGAAGAAGTCATTGTTTTTTGCTTAGAGAAACTGGAAGCAGTTTCTTAAAGGTTGTGGTTTTTTGGTTTTTTTTAACAGGCAGAAGACAATTTGCGTAGAGAAAAAAACCTGGAAGAAGCAAAGAAGATTACCATTAAAAAAGATCCAAATCTTCAAGAGCCAGAATGTGTGAGTAAAAGCTTGATGAACTTCTCCATCTTTAAGAGGTGGGGTTGGATTTGTGCAGTAGACCGTCTCTGCAAAACAAAAACACCTCATCTCAGCTATTCTTCAGGCAAGCCCAAATAGTTC ........................................................................................................................................................................................................((((((..((.(((((.((((.......)).)).....))))).))....(((((((((...))))))))).))))))..(((((.((.....))))))).................................................. ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|