| (1) HEART | (1) KIDNEY | (1) OTHER |
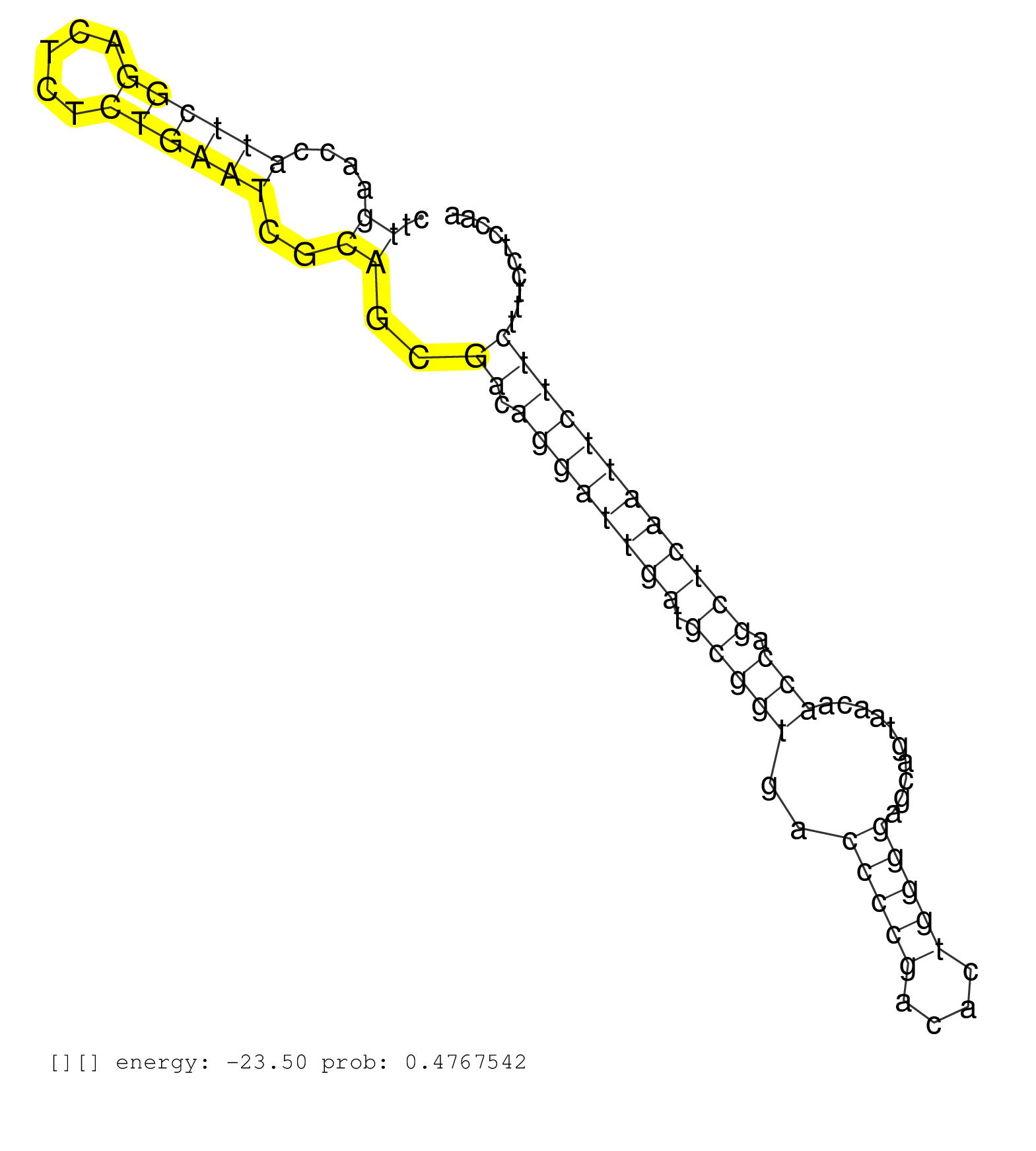
| CATTGGTGAATCCCTCCCCCAATGACTGTTTTCGAGAATGACAAAACGGACTCAGTAGTCAGATGTACGATTGGTTACGTTTCCTTCCGTGGGAAATGAAAATCTCATTACCTTGTTTTCCGTTGCCACAATTAATTTTGCAATCATCTGGATTTCCTCCTTTTTATAGCGGAAAACAAGTCCCCTCCTCGTCCCTGTGGCTTGAACCATTCGGACTCTCTGAATCGCAGCGACAGGATTGATGCGGTGACCCCGACACTGGGGAGCAGTAACAACCAGCTCAATTCTTCTTTCCTCCAAGTGTACATCCCTGACTACTCAGTGAGAGCCCTCTCAGACATTCAGTTTGT ..........................................................................................................................................................................................................((....((((((.....))))))..))..((.((((((((.(((((..(((((....)))))..........))).))))))))))))............................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TTCGGACTCTCTGAATCGCAGCGACAGG................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 4.00 | - |
| ....................................................................................................................................................................................................................GGACTCTCTGAATCGC.......................................................................................................................... | 16 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 |
| ....................AATGACTGTTTTCGAGAATGACA................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .................................................................................................................................................................................................................TTCGGACTCTCTGAATCGCAGCGACAG.................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| .....................................................................................................................................................................................................................GACTCTCTGAATCGCAGCGACAGGATTGATG.......................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ...........................................AAACGGACTCAGTAG.................................................................................................................................................................................................................................................................................................... | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ................................................................................................................................................................................................................ATTCGGACTCTCTGAATCGCAGCGACA................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................................................................................GGACTCTCTGAATCGCAGCG...................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ................................................................................................................................................................................................................ATTCGGACTCTCTGAATCGCAGCGACAGG................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - |
| ...............................................................................................................................................................................................................CATTCGGACTCTCTGAATCGCAG........................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 |
| ....................................................................................................................................................................................................................................AGCGACAGGATTGAT........................................................................................................... | 15 | 2 | 0.50 | 0.50 | 0.50 | - | - |
| .......................................................................................................................................................................................................................................GACAGGATTGATGCG........................................................................................................ | 15 | 2 | 0.50 | 0.50 | 0.50 | - | - |
| .....................................................................................................................................................................................................................GACTCTCTGAATCGC.......................................................................................................................... | 15 | 3 | 0.33 | 0.33 | 0.33 | - | - |
| CATTGGTGAATCCCTCCCCCAATGACTGTTTTCGAGAATGACAAAACGGACTCAGTAGTCAGATGTACGATTGGTTACGTTTCCTTCCGTGGGAAATGAAAATCTCATTACCTTGTTTTCCGTTGCCACAATTAATTTTGCAATCATCTGGATTTCCTCCTTTTTATAGCGGAAAACAAGTCCCCTCCTCGTCCCTGTGGCTTGAACCATTCGGACTCTCTGAATCGCAGCGACAGGATTGATGCGGTGACCCCGACACTGGGGAGCAGTAACAACCAGCTCAATTCTTCTTTCCTCCAAGTGTACATCCCTGACTACTCAGTGAGAGCCCTCTCAGACATTCAGTTTGT ..........................................................................................................................................................................................................((....((((((.....))))))..))..((.((((((((.(((((..(((((....)))))..........))).))))))))))))............................................................ ........................................................................................................................................................................................................201................................................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|