| (1) BRAIN | (1) HEART | (1) KIDNEY | (2) OTHER |
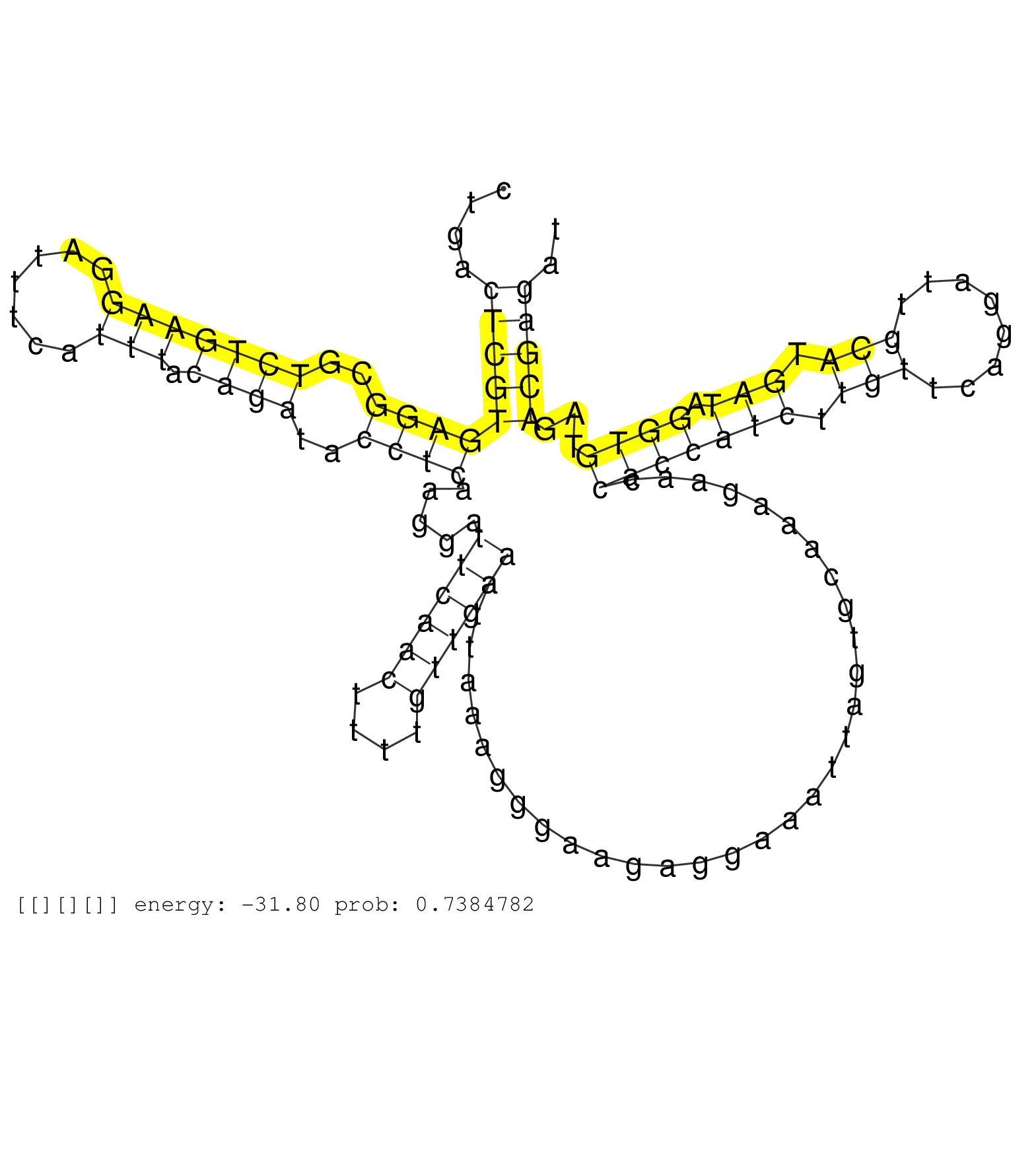
| TACAAAAAACACACACACACAAAAAAACTATTCCAGGTGGACCAAGAATTCACAATTTAATAACCTAACCTGAAAAATAATAAAGAGAATCCTGACTCGTGAGGCGTCTGAAGGATTTCATTTACAGATACCTCAAGGATTCAACTTTTGTTGAAATTAAAGGGAAGAGGAAATTAGTGCAAAGAACCACCATCTTGTTCAGGATTGCATGATAGGTGTAGACGAGATATCTTGAGAGCATTGCAATTCGGTGCAAGATGTTTGTTAAAGAGCCATTTTGGCTTCTTAAAAACAAAATTTAAAAAAATGGACTCTTTGCTTCAACATCACCCCTTCCCATCATAATGCTCATAATAACACTTTAGGGG ...............................................................................................(((((((((..(((((((.......))).))))..)))).....((((((....))))))................................(((((((.(((........))).))).))))...))))).............................................................................................................................................. ...........................................................................................92......................................................................................................................................228.......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553592(SRX182798) source: Brain. (Brain) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................AGGTGTAGACGAGATATCTTGAGAGC................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ...................................................................................................................................................................................................TGTTCAGGATTGCATGATAGGTGTAGAC................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ................................................................................................TCGTGAGGCGTCTGAAGGA............................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - |
| ..................................................................................AAGAGAATCCTGACTCGTGAGGCGTC.................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - |
| .....................................................................................AGAATCCTGACTCGTGAGGCGTCTGA................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................CATGATAGGTGTAGAAT................................................................................................................................................ | 17 | 1.00 | 0.50 | - | - | - | - | 1.00 | |
| ...............................................................................................................................................................................................................CATGATAGGTGTAGAATAA.............................................................................................................................................. | 19 | 1.00 | 0.50 | - | 1.00 | - | - | - | |
| ...............................................................................................................................................................................................................CATGATAGGTGTAGAATA............................................................................................................................................... | 18 | 1.00 | 0.50 | - | - | 1.00 | - | - | |
| ...............................................................................................................................................................................................................CATGATAGGTGTAGA.................................................................................................................................................. | 15 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - |
| TACAAAAAACACACACACACAAAAAAACTATTCCAGGTGGACCAAGAATTCACAATTTAATAACCTAACCTGAAAAATAATAAAGAGAATCCTGACTCGTGAGGCGTCTGAAGGATTTCATTTACAGATACCTCAAGGATTCAACTTTTGTTGAAATTAAAGGGAAGAGGAAATTAGTGCAAAGAACCACCATCTTGTTCAGGATTGCATGATAGGTGTAGACGAGATATCTTGAGAGCATTGCAATTCGGTGCAAGATGTTTGTTAAAGAGCCATTTTGGCTTCTTAAAAACAAAATTTAAAAAAATGGACTCTTTGCTTCAACATCACCCCTTCCCATCATAATGCTCATAATAACACTTTAGGGG ...............................................................................................(((((((((..(((((((.......))).))))..)))).....((((((....))))))................................(((((((.(((........))).))).))))...))))).............................................................................................................................................. ...........................................................................................92......................................................................................................................................228.......................................................................................................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553592(SRX182798) source: Brain. (Brain) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) |
|---|