| (1) BRAIN | (1) HEART | (1) KIDNEY | (1) OTHER |
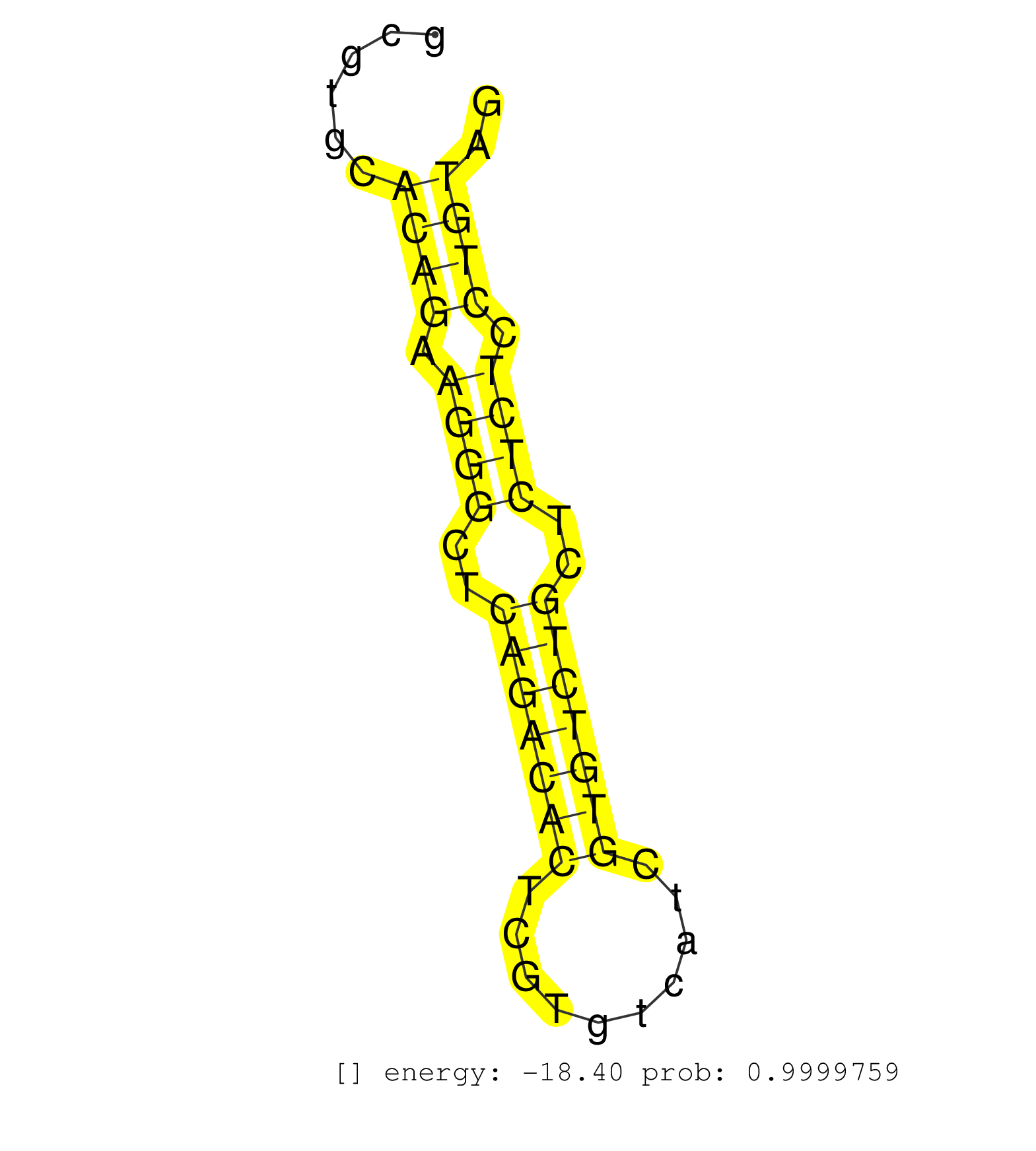
| GCAGTCAAGTGGCAGAGTCAGGATTAGAACCCGGATCCTCTGAATCCCAAGGCCGTGCTCTTTGAACTAGGCCGTGCTGCTTCTCTGCAGTTGCTTCGCTGTAGCGTCATCATCATCCTCAGTGGTATTTATTGAGTGCTTTCTATATATCCGAAGCCCTCGGGAGGGTACAATATAACGGAATTAGCAGTCACGTTGCCTGCCCGTAACGAGCTTTCAGTCTCGAGTCTTCACAGCGGAGGGATCGCGTGCACAGAAGGGCTCAGACACTCGTGTCATCGTGTCTGCTCTCTCCTGTAGGTTAATGAAGGCCAGCCTGGAAGGAGACCTCATCAGGACCCAGAAGCTGG ............................................................................................................................................................................................................................................................((((.((((..(((((((..........)))))))..)))).)))).................................................... ......................................................................................................................................................................................................................................................247..................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTAG.................................................. | 21 | 1 | 9.00 | 9.00 | 7.00 | 2.00 | - | - |
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTAGT................................................. | 22 | 6.00 | 9.00 | 6.00 | - | - | - | |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACACTCGA............................................................................ | 23 | 6.00 | 5.00 | 5.00 | - | - | 1.00 | |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACACTCGT............................................................................ | 23 | 1 | 5.00 | 5.00 | 5.00 | - | - | - |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACACTCG............................................................................. | 22 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | 1.00 |
| .................................................................................................................................................................................................................................................................AGGGCTCAGACACTCG............................................................................. | 16 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTAGTA................................................ | 23 | 3.00 | 9.00 | 3.00 | - | - | - | |
| .........................................................................................................................................................................................................................................................................................TGTCTGCTCTCTCCTGTAGATA............................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | |
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTAGTT................................................ | 23 | 2.00 | 9.00 | 2.00 | - | - | - | |
| ..........................................................................................................................................................................................................................................................................................................................................CATCAGGACCCAGAAGCT.. | 18 | 1 | 2.00 | 2.00 | - | - | 2.00 | - |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACACAT.............................................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| ........................................................................................................................................................................................................................................................................................................................................CTCATCAGGACCCAGAAGCTG. | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTAGTATT.............................................. | 25 | 1.00 | 9.00 | 1.00 | - | - | - | |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACAA................................................................................ | 19 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................................................................................................................................................................................................TGTCTGCTCTCTCCTGTAGTA................................................ | 21 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................................................................................................................................................................................................TGTCTGCTCTCTCCTGTAGA................................................. | 20 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .........................................................................................................................................................................................................................................................TGCACAGAAGGGCTCAGACACA............................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .......................................................................................................................................................................................................................................................................................CGTGTCTGCTCTCTCCTGTATTA................................................ | 23 | 1.00 | 0.00 | 1.00 | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................TGGAAGGAGACCTCATCAGGA............ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................................................................CACAGAAGGGCTCAGACACTA.............................................................................. | 21 | 1.00 | 0.00 | 1.00 | - | - | - |
| GCAGTCAAGTGGCAGAGTCAGGATTAGAACCCGGATCCTCTGAATCCCAAGGCCGTGCTCTTTGAACTAGGCCGTGCTGCTTCTCTGCAGTTGCTTCGCTGTAGCGTCATCATCATCCTCAGTGGTATTTATTGAGTGCTTTCTATATATCCGAAGCCCTCGGGAGGGTACAATATAACGGAATTAGCAGTCACGTTGCCTGCCCGTAACGAGCTTTCAGTCTCGAGTCTTCACAGCGGAGGGATCGCGTGCACAGAAGGGCTCAGACACTCGTGTCATCGTGTCTGCTCTCTCCTGTAGGTTAATGAAGGCCAGCCTGGAAGGAGACCTCATCAGGACCCAGAAGCTGG ............................................................................................................................................................................................................................................................((((.((((..(((((((..........)))))))..)))).)))).................................................... ......................................................................................................................................................................................................................................................247..................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|