| (1) BRAIN | (1) HEART | (1) KIDNEY | (1) OTHER |
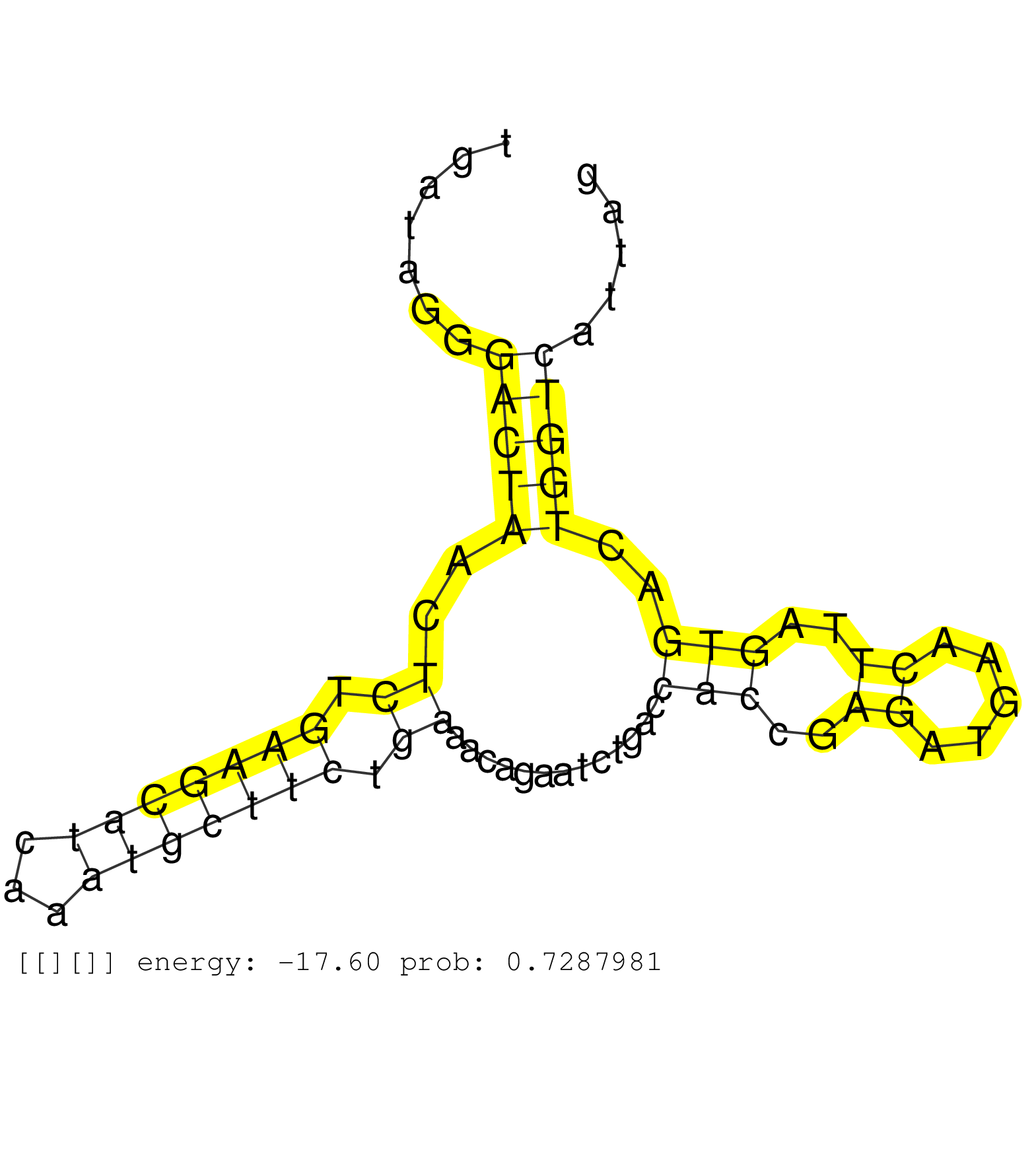
| ACTCTCAAGCTTGCGCTCTGTCCACTAGGCCTCGCTGCTTCTCAGTCTGTTAAAACCTGAATTTGAAACTGTAATGTTTTGGTACAAAAGTAAGAATCTGTGTTCTGTTAACCTGGATACAAGTTTTCTGTTCAATTCTGGAAAAAAAAAAATTATCTTTGTGTTAACTTTATGCCTTAGAAAAATGAAATCAGCCAATTATGTATAACTTTTAACTCTTGATAGGGACTAACTCTGAAGCATCAAATGCTTCTGAAACAGAATCTGACCACCGAGATGAACTTAGTGACTGGTCATTAGCCCCAGCAGAGGAAGAAAGGGACAATTACCTACGCAGAGGAGATGGGCGA ..................................................................................................................................................................................................................................(((((..((.(((((((...))))))).)).............(((..((.....))..)))..)))))....................................................... ...........................................................................................................................................................................................................................220.............................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................GGGACTAACTCTGAAGC............................................................................................................. | 17 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................TCTGAAACAGAATCTGACCACC............................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................GAGATGAACTTAGTGACTGGT........................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..........................................................GAATTTGAAACTGTAGAC.................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | |
| ...........................................................................................................................................................................................................................................................TCTGAAACAGAATCTGACCACCGAGATGAACT................................................................... | 32 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................................TAACTCTGAAGCATCAAATGC.................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .....................................................................................................................................................................................................................................................AATGCTTCTGAAACAGAATCTGACCACCGAG.......................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................GAGATGAACTTAGTGACTGGTCATTAG.................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - |
| .....................................................................................................................................................................................................................................................AATGCTTCTGAAACAGAATCTGACCACCG............................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................ACTAACTCTGAAGCATCAAATGC.................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................................TAGTGACTGGTCATTAGCCCCAGCAGAGGA..................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ................................................................................................................................................................................................................................GGGACTAACTCTGAAG.............................................................................................................. | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - |
| .........................................................................................................................................................................................................................................TCTGAAGCATCAAAT...................................................................................................... | 15 | 13 | 0.08 | 0.08 | - | 0.08 | - | - |
| ACTCTCAAGCTTGCGCTCTGTCCACTAGGCCTCGCTGCTTCTCAGTCTGTTAAAACCTGAATTTGAAACTGTAATGTTTTGGTACAAAAGTAAGAATCTGTGTTCTGTTAACCTGGATACAAGTTTTCTGTTCAATTCTGGAAAAAAAAAAATTATCTTTGTGTTAACTTTATGCCTTAGAAAAATGAAATCAGCCAATTATGTATAACTTTTAACTCTTGATAGGGACTAACTCTGAAGCATCAAATGCTTCTGAAACAGAATCTGACCACCGAGATGAACTTAGTGACTGGTCATTAGCCCCAGCAGAGGAAGAAAGGGACAATTACCTACGCAGAGGAGATGGGCGA ..................................................................................................................................................................................................................................(((((..((.(((((((...))))))).)).............(((..((.....))..)))..)))))....................................................... ...........................................................................................................................................................................................................................220.............................................................................300................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553595(SRX182801) source: Kidney. (Kidney) | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................................................................AGTTCATCTCGGTGG................................................................... | 15 | 3 | 13.67 | 13.67 | 13.00 | 0.33 | - | 0.33 |
| ............................................................................................................................................................................................................................................................................aAGTTCATCTCGGTGA.................................................................. | 16 | 5.67 | 0.00 | 5.33 | 0.33 | - | - | |
| ............................................................................................................................................................................................................................................................................ccaaAGTTCATCTCGAACC............................................................... | 19 | 2.33 | 0.00 | 1.67 | 0.67 | - | - | |
| ............................................................................................................................................................................................................................................................................caaAGTTCATCTCGGAAC................................................................ | 18 | 2.00 | 0.00 | 1.67 | 0.33 | - | - | |
| ............................................................................................................................................................................................................................................................................cagAGTTCATCTCGGGAC................................................................ | 18 | 0.67 | 0.00 | 0.33 | - | - | 0.33 | |
| ............................................................................................................................................................................................................................................................................aaAGTTCATCTCGGTAA................................................................. | 17 | 0.33 | 0.00 | - | 0.33 | - | - | |
| ............................................................................................................................................................................................................................................................................ccagAGTTCATCTCGGACC............................................................... | 19 | 0.33 | 0.00 | - | 0.33 | - | - |