| (1) BRAIN | (1) HEART | (2) OTHER |
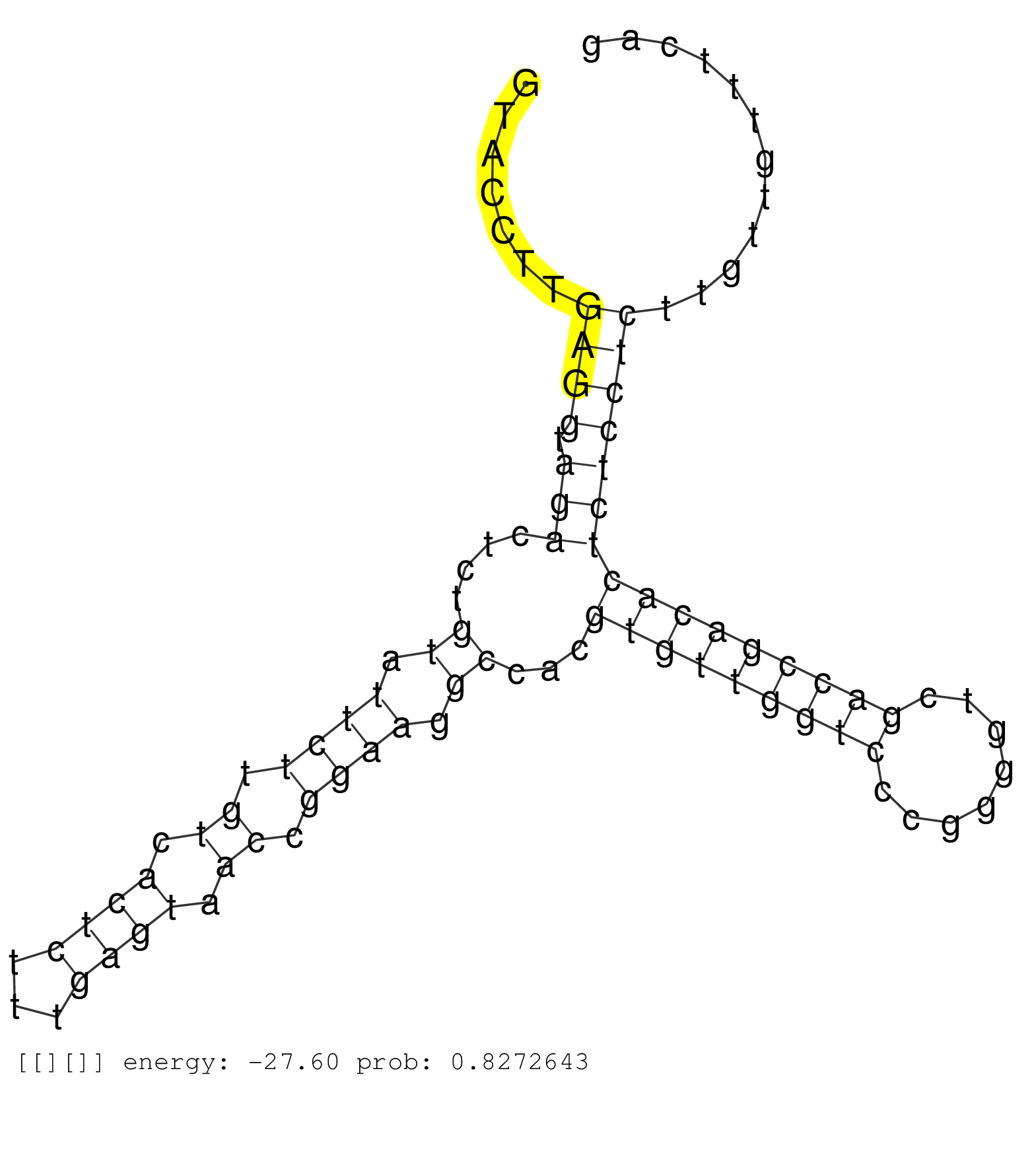
| GCAGTTGACATCTGTTCCAGCAGCCCTGCATGGAAGCCATTGCTTAAAAAGTAGGAATTCTGCTACTTTTAGCATGTTTGTCACTGATTACATGCAGTAATGGTTCTTTAATGAGAAGATAAAAGATTGGCCATTTTTCAAATGAAAACCAGACCCTCTGTCTGCATTTCACGAATGAGCTGCCCTAGTGAGATCTCAACCTGTCAAATCAGTCGCTTCCAAAGCCAGCTCAGCTTCCGTTGTATTTACCGTGGAAATGCGACTTCCCCAAGACGCTTCTGTCACTGTCACAGATTGAGCCTGTCCCTTCACTTCGCCCCCCCGCTCTCGAGTCCAGCTGTGTACCTTGAGGTAGACTCTGTATTCTTGTCACTCTTTGAGTAACCGGAAGGCCACGTGTTGGTCCCGGGGTCGACCGACACTCTCCTCTTGTTGTTTCAGGATGAACACAGTACAGTGTAGAGCGCTCACGTGCCTTCAGAGTATTGTGT ............................................................................................................................................................................................................................................................................................................................................................((((.(((....((.((((.((.((((...)))).)).)))).))...(((((((((........)))))))))))))))).............................................................. .....................................................................................................................................................................................................................................................................................................................................................342................................................................................................441................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................................................................CAGCTGTGTACCTTGAG............................................................................................................................................ | 17 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - |
| ..........................................................................................................................................................................................................GTCAAATCAGTCGCT.................................................................................................................................................................................................................................................................................. | 15 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................ACCGGAAGGCCACGTGTTGGTCCCGGGGTCGACC.......................................................................... | 34 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| ...GTTGACATCTGTTCCAGC...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - |
| .........................................................................................................................................................................CACGAATGAGCTGCC................................................................................................................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | 0.50 | - | - |
| GCAGTTGACATCTGTTCCAGCAGCCCTGCATGGAAGCCATTGCTTAAAAAGTAGGAATTCTGCTACTTTTAGCATGTTTGTCACTGATTACATGCAGTAATGGTTCTTTAATGAGAAGATAAAAGATTGGCCATTTTTCAAATGAAAACCAGACCCTCTGTCTGCATTTCACGAATGAGCTGCCCTAGTGAGATCTCAACCTGTCAAATCAGTCGCTTCCAAAGCCAGCTCAGCTTCCGTTGTATTTACCGTGGAAATGCGACTTCCCCAAGACGCTTCTGTCACTGTCACAGATTGAGCCTGTCCCTTCACTTCGCCCCCCCGCTCTCGAGTCCAGCTGTGTACCTTGAGGTAGACTCTGTATTCTTGTCACTCTTTGAGTAACCGGAAGGCCACGTGTTGGTCCCGGGGTCGACCGACACTCTCCTCTTGTTGTTTCAGGATGAACACAGTACAGTGTAGAGCGCTCACGTGCCTTCAGAGTATTGTGT ............................................................................................................................................................................................................................................................................................................................................................((((.(((....((.((((.((.((((...)))).)).)))).))...(((((((((........)))))))))))))))).............................................................. .....................................................................................................................................................................................................................................................................................................................................................342................................................................................................441................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR553594(SRX182800) source: Heart. (Heart) | SRR553593(SRX182799) source: Cerebellum. (Cerebellum) | SRR553592(SRX182798) source: Brain. (Brain) | SRR553596(SRX182802) source: Testis. (Testis) |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................................................................................................................................................................................acTCGGTCGACCCCGCA...................................................................... | 17 | 1.00 | 0.00 | - | - | - | 1.00 |