| (1) KIDNEY | (1) OTHER |
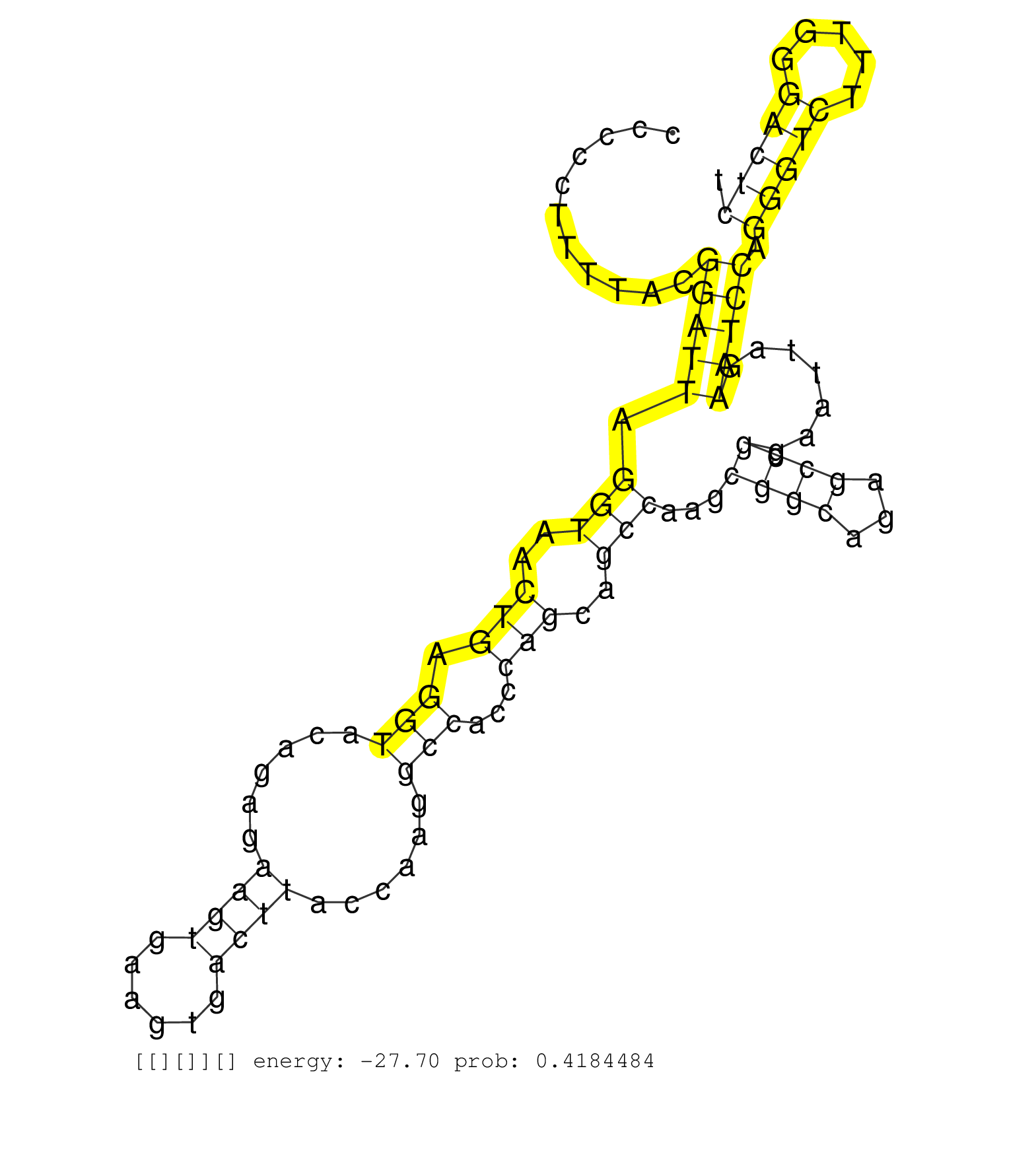
| TGTCCCCTCCCAAGCGCTCAGGACGGTGCTCCGTTTAGAGTAAGCGCTCAATAAATGCGGTCCCTTGATGACTGTAATAATAATATTTAAGTGCTTACTACGCGTAAAGCACTAAGCCCTAGGGTAGGTACGAGCTAATCAGTTCGGACAGTCCCTACCCCACATGGGAGTCTTAATCCCCCTTTTACGGATTAGGTAACTGAGGTACAGAGAAGTGAAGTGACTTACCAAGGCCACCCAGCAGCCAAGCGGCAGAGCCGGAATTAGAATCCAGGGTCTTTGGGACTCTTTTGCTTTTAGCTCCGCAAAAGCAGCCCAACCTCCCCAACCCCTGCCTGCGTCAACAGCTG ............................................................................................................................................................................................(((((.(((..(((.(((......((((......))))......)))...)))..)))...((((...)))).......))))).(((((.....))))).............................................................. .................................................................................................................................................................................178............................................................................................................289........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................TTTTACGGATTAGGTAACTGAAGC................................................................................................................................................ | 24 | 4.00 | 0.00 | 4.00 | - | |
| ......................................................................................................................................................................................TTTTACGGATTAGGTAACTGAAGCA............................................................................................................................................... | 25 | 4.00 | 0.00 | 4.00 | - | |
| ......................................................................................................................................................................................TTTTACGGATTAGGTAACTGAAG................................................................................................................................................. | 23 | 4.00 | 0.00 | 4.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAGCA............................................................................................................................................... | 24 | 3.00 | 0.00 | 3.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAGC................................................................................................................................................ | 23 | 3.00 | 0.00 | 3.00 | - | |
| .........................................................................................................................................................................................TACGGATTAGGTAACTGAAGC................................................................................................................................................ | 21 | 2.00 | 0.00 | 2.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAGCT............................................................................................................................................... | 24 | 2.00 | 0.00 | 2.00 | - | |
| ........................................................................................................................................................................................TTACGGATTAGGTAACTGAAGCA............................................................................................................................................... | 23 | 2.00 | 0.00 | 2.00 | - | |
| .........................................................................................................................................................................................TACGGATTAGGTAACTGAAA................................................................................................................................................. | 20 | 1.00 | 0.00 | 1.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAG................................................................................................................................................. | 22 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................................................................................................TACGGATTAGGTAACTGAAGCA............................................................................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | |
| ........................................................................................................................................................................................TTACGGATTAGGTAACTGAAGA................................................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | |
| ........................................................................................................................................................................................TTACGGATTAGGTAACTGAAGC................................................................................................................................................ | 22 | 1.00 | 0.00 | 1.00 | - | |
| ..........................................................................................................................................................................................................................................................................GAATCCAGGGTCTTTGGGA................................................................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAGA................................................................................................................................................ | 23 | 1.00 | 0.00 | 1.00 | - | |
| .........................................................................................................................................................................................TACGGATTAGGTAACTGAAAA................................................................................................................................................ | 21 | 1.00 | 0.00 | 1.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAAA................................................................................................................................................. | 22 | 1.00 | 0.00 | 1.00 | - | |
| .......................................................................................................................................................................................TTTACGGATTAGGTAACTGAA.................................................................................................................................................. | 21 | 1.00 | 0.00 | 1.00 | - |
| TGTCCCCTCCCAAGCGCTCAGGACGGTGCTCCGTTTAGAGTAAGCGCTCAATAAATGCGGTCCCTTGATGACTGTAATAATAATATTTAAGTGCTTACTACGCGTAAAGCACTAAGCCCTAGGGTAGGTACGAGCTAATCAGTTCGGACAGTCCCTACCCCACATGGGAGTCTTAATCCCCCTTTTACGGATTAGGTAACTGAGGTACAGAGAAGTGAAGTGACTTACCAAGGCCACCCAGCAGCCAAGCGGCAGAGCCGGAATTAGAATCCAGGGTCTTTGGGACTCTTTTGCTTTTAGCTCCGCAAAAGCAGCCCAACCTCCCCAACCCCTGCCTGCGTCAACAGCTG ............................................................................................................................................................................................(((((.(((..(((.(((......((((......))))......)))...)))..)))...((((...)))).......))))).(((((.....))))).............................................................. .................................................................................................................................................................................178............................................................................................................289........................................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR553596(SRX182802) source: Testis. (Testis) | SRR553595(SRX182801) source: Kidney. (Kidney) |
|---|---|---|---|---|---|---|
| ........................................................................................................................................................................atgCGTAAAAGGGGGATTAAGGTA.............................................................................................................................................................. | 24 | 1.00 | 0.00 | 1.00 | - | |
| ........................................................TATTATTATTACAGTCATCAAGGGACCGC......................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - |