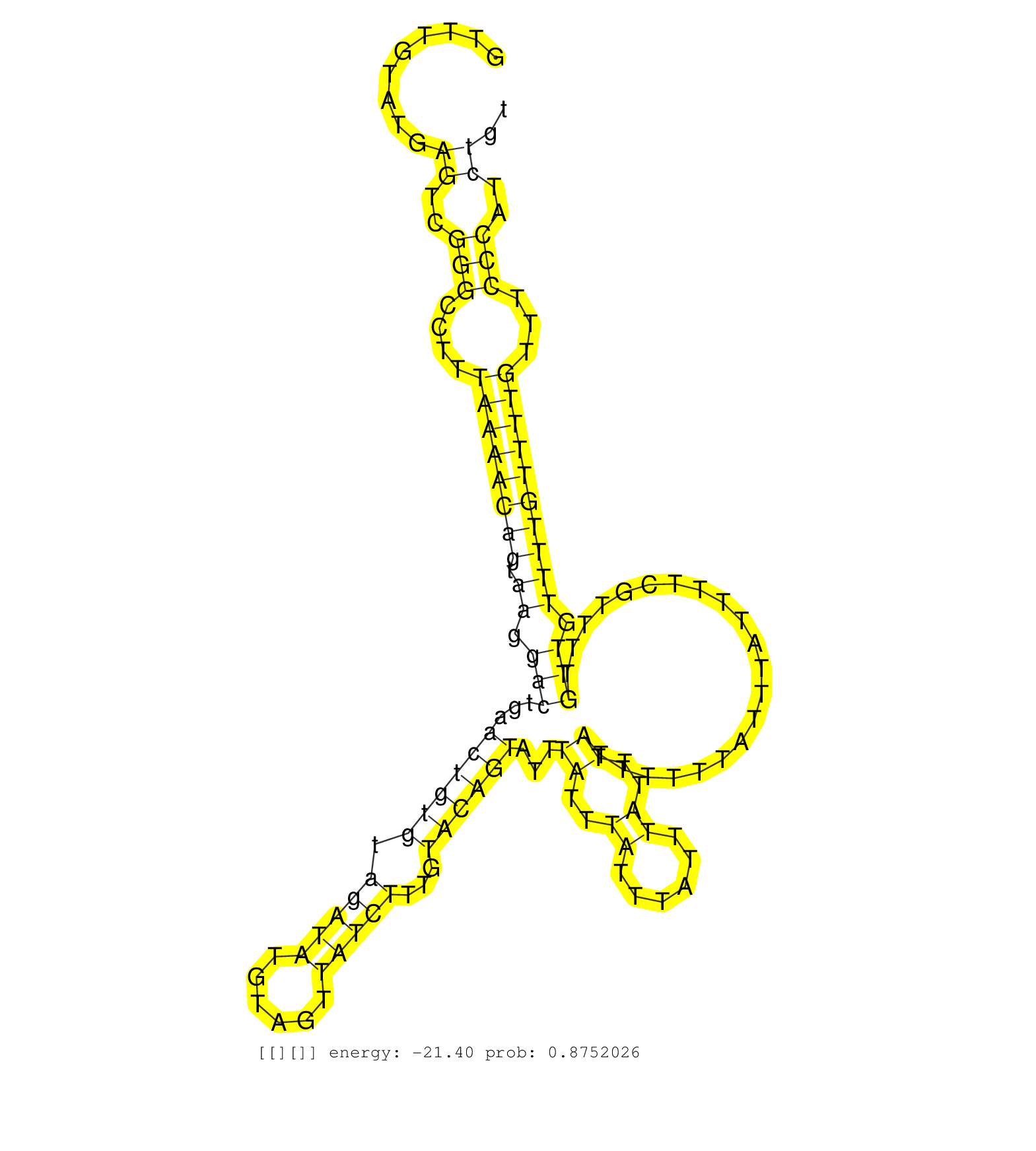

| Gene: Pramel3 | ID: uc009uhd.1_intron_1_0_chrX_131837303_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| GGCACGAGGTTTTGAAGTTCTACGTCAACCTACATTAACGTGGTGAGAAGGTTTGTATGAGTCGGGCCTTTAAAACAGTAAGGACTGAACTGTGTAGATATGTAGTTATCTTTGTACAGTATTTATTTATTTATTTATTTATTTTTTATTTATTTTCGTTTTGTTGTTTTGTTTTGTTTCCCATCTGTCAGTTCATGTTATTTTCTGAGCTGTTTAGACACCAAGGGTCCCGGTTTAGAAAGGCTTTAAT ...........................................................((..(((....((((((((.((.(((...((((((.(((((......)))))...))))))...((..((......))..)).....................))).))))))))))...)))..))................................................................ ..................................................51.......................................................................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................ATATGTAGTTATCTTTGTACAGTATTTATTTATTTATTTATTTATTTTTTAT..................................................................................................... | 52 | 1 | 67.00 | 67.00 | 67.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TACATTAACGTGGTGAGAAGGTTcatt................................................................................................................................................................................................. | 27 | catt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TTGAAGTTCTACGTCAACCTACATTAAC................................................................................................................................................................................................................... | 28 | 3 | 1.00 | 1.00 | - | - | 0.33 | - | - | - | - | - | - | 0.33 | - | 0.33 | - | - | - |
| ..........................................................................................TGTGTAGATATGTAGTaaag............................................................................................................................................ | 20 | aaag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................TACATTAACGTGGTGAGAAGGTTc.................................................................................................................................................................................................... | 24 | c | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TAACGTGGTGAGAAGGTTTGTATGA.............................................................................................................................................................................................. | 25 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - |
| ............TGAAGTTCTACGTCAACCTACATTA..................................................................................................................................................................................................................... | 25 | 5 | 0.60 | 0.60 | - | - | 0.60 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TTGAAGTTCTACGTCAACCTACATTA..................................................................................................................................................................................................................... | 26 | 5 | 0.40 | 0.40 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | 0.20 |
| ....CGAGGTTTTGAAGTTCTACGTCAACCTA.......................................................................................................................................................................................................................... | 28 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - |
| ..CACGAGGTTTTGAAGTTCTACGTCAAC............................................................................................................................................................................................................................. | 27 | 5 | 0.40 | 0.40 | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | 0.20 | - |
| ..................................................GTTTGTATGAGTCGGGCCTTTAAAAC.............................................................................................................................................................................. | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ...........................................TGAGAAGGTTTGTATGAGTCGGGCCT..................................................................................................................................................................................... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ..........................................GTGAGAAGGTTTGTATGAGTCGGG........................................................................................................................................................................................ | 24 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................AAGGACTGAACTGTGTAGATATGTAGTTATC............................................................................................................................................ | 31 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| .................TTCTACGTCAACCTACATTAACGTGGTG............................................................................................................................................................................................................. | 28 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GTGAGAAGGTTTGTATGAGTCGGGC....................................................................................................................................................................................... | 25 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ..........TTTGAAGTTCTACGTCAACCTACATTAA.................................................................................................................................................................................................................... | 28 | 5 | 0.20 | 0.20 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - |
| GGCACGAGGTTTTGAAGTTCTACGTCAACCTACATTAACGTGGTGAGAAGGTTTGTATGAGTCGGGCCTTTAAAACAGTAAGGACTGAACTGTGTAGATATGTAGTTATCTTTGTACAGTATTTATTTATTTATTTATTTATTTTTTATTTATTTTCGTTTTGTTGTTTTGTTTTGTTTCCCATCTGTCAGTTCATGTTATTTTCTGAGCTGTTTAGACACCAAGGGTCCCGGTTTAGAAAGGCTTTAAT ...........................................................((..(((....((((((((.((.(((...((((((.(((((......)))))...))))))...((..((......))..)).....................))).))))))))))...)))..))................................................................ ..................................................51.......................................................................................................................................188............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|