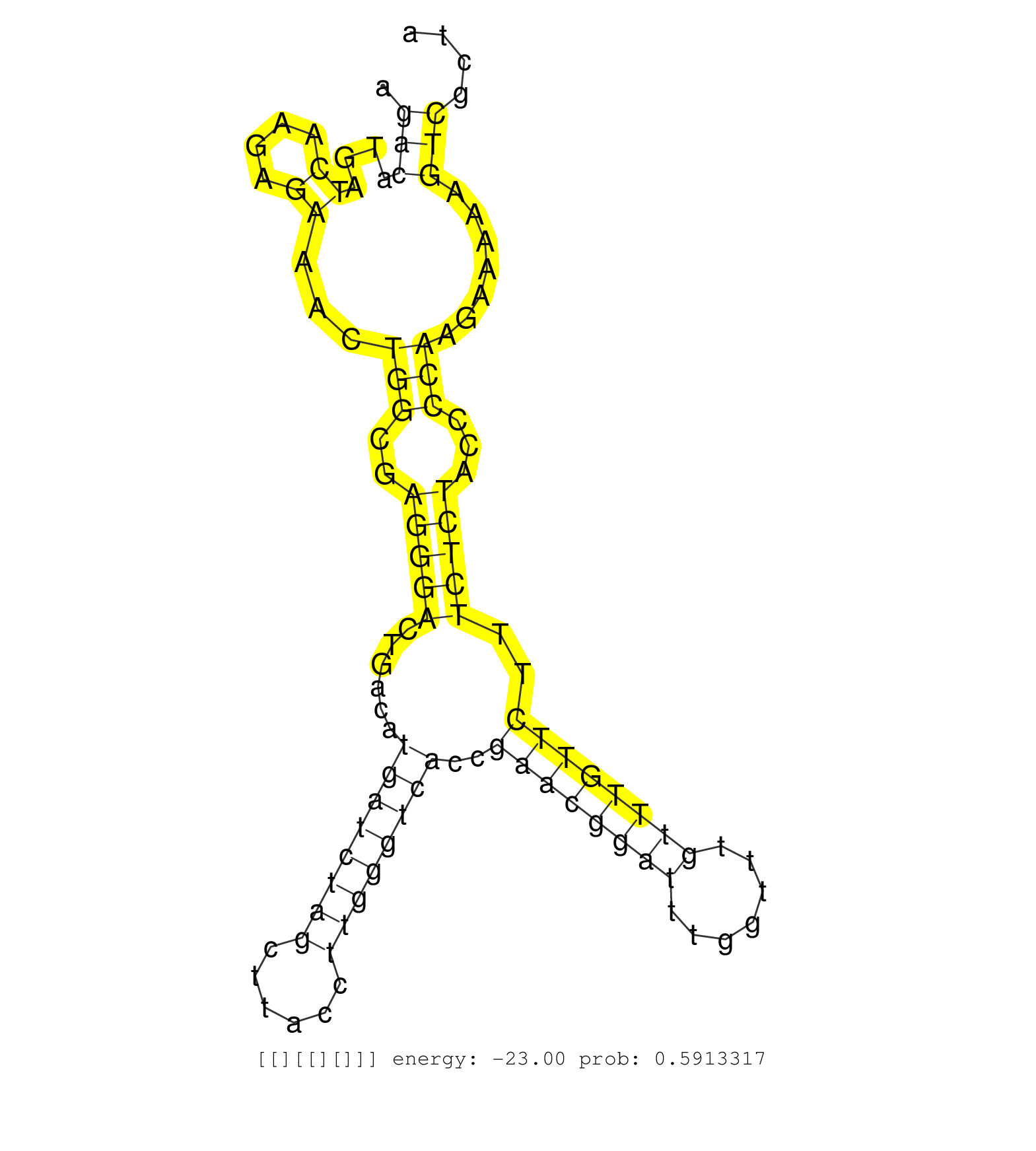

| Gene: AK086455 | ID: uc009uco.1_intron_0_0_chrX_106030025_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| GAGCTCCGGAGTTGCTTACCCTTCCTGTAAGGGCAATCGGACCCTTTCCGGTGAGGGAACCTCTCTCCAGACATGATCAAGAGAAACTGGCGAGGGACTGACATGATCTAGCTTACCTTGGGTCACCGAACGGATTTGGTTTGTTTGTTCTTTCTCTACCCCAAGAAAAAGTCGCTAGCTACTGCAAGGTTTTGTTTTCAAACAAGGAACAGCATGCCAAAAGAAAGTGCTGCGCCTACCTGATGAGCAA .....................................................................(((....((....))...(((..(((((......((((((((......))))))))..((((((((.......))))))))..)))))...))).......)))............................................................................. ....................................................................69..........................................................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TGATCAAGAGAAACTGGCGAGGGACTG...................................................................................................................................................... | 27 | 1 | 12.00 | 12.00 | 5.00 | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| .........................................................................TGATCAAGAGAAACTGGCGAGGGACT....................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................AGGGACTGACATGATatca........................................................................................................................................... | 19 | atca | 4.00 | 0.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................AGGGACTGACATGATCTA............................................................................................................................................ | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGGGTCACCGAACGGATTTGGcatg........................................................................................................... | 25 | catg | 3.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GGGACTGACATGATCTAG........................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GAAAGTGCTGCGCCTACCTGATGAGC.. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGATCAAGAGAAACTGGCGAGGGAC........................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGACATGATCTAGCTTACCTTGGGTC.............................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| .................................................................................................................TACCTTGGGTCACCGAACGGATTTGGcat............................................................................................................ | 29 | cat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................ATGATCTAGCTTACCTTG.................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................CAAGAGAAACTGGCGAGGGACTGACATGA................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGATCAAGAGAAACTGGCGAGGGACTGA..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGATCAAGAGAAACTGGCGAGGGAatg...................................................................................................................................................... | 27 | atg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGCTTACCTTGGGTCACCGAACGGATT.................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGCCAAAAGAAAGTGCTGCGCCTACCTGA....... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TCCAGACATGATCAAGAGAAACTGGCG.............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGGGTCACCGAACGGATTTGGTTTGT.......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................AGAGAAACTGGCGAGGGA......................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGCGAGGGACTGACATGAT............................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................ATCAAGAGAAACTGGCGAGGGACTG...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................AGACATGATCAAGAGAAACTGGCGAGG........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TTGTTCTTTCTCTACCCCAAGAAAAAGTC............................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TACCTTGGGTCACCGAACGGATTTGGca............................................................................................................. | 28 | ca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TAGCTTACCTTGGGTCACCGAACGG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGACATGATCTAGCTTACCTTGGGTCA............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GAGCTCCGGAGTTGCTTACCCTTCCTGTAAGGGCAATCGGACCCTTTCCGGTGAGGGAACCTCTCTCCAGACATGATCAAGAGAAACTGGCGAGGGACTGACATGATCTAGCTTACCTTGGGTCACCGAACGGATTTGGTTTGTTTGTTCTTTCTCTACCCCAAGAAAAAGTCGCTAGCTACTGCAAGGTTTTGTTTTCAAACAAGGAACAGCATGCCAAAAGAAAGTGCTGCGCCTACCTGATGAGCAA .....................................................................(((....((....))...(((..(((((......((((((((......))))))))..((((((((.......))))))))..)))))...))).......)))............................................................................. ....................................................................69..........................................................................................................177....................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TTGTTCTTTCTCTACCCC........................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGTTTTCAAACAAGGAACAGCATGCCA............................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCCGGAGTTGCTTACCCTTCCTGTAA............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |