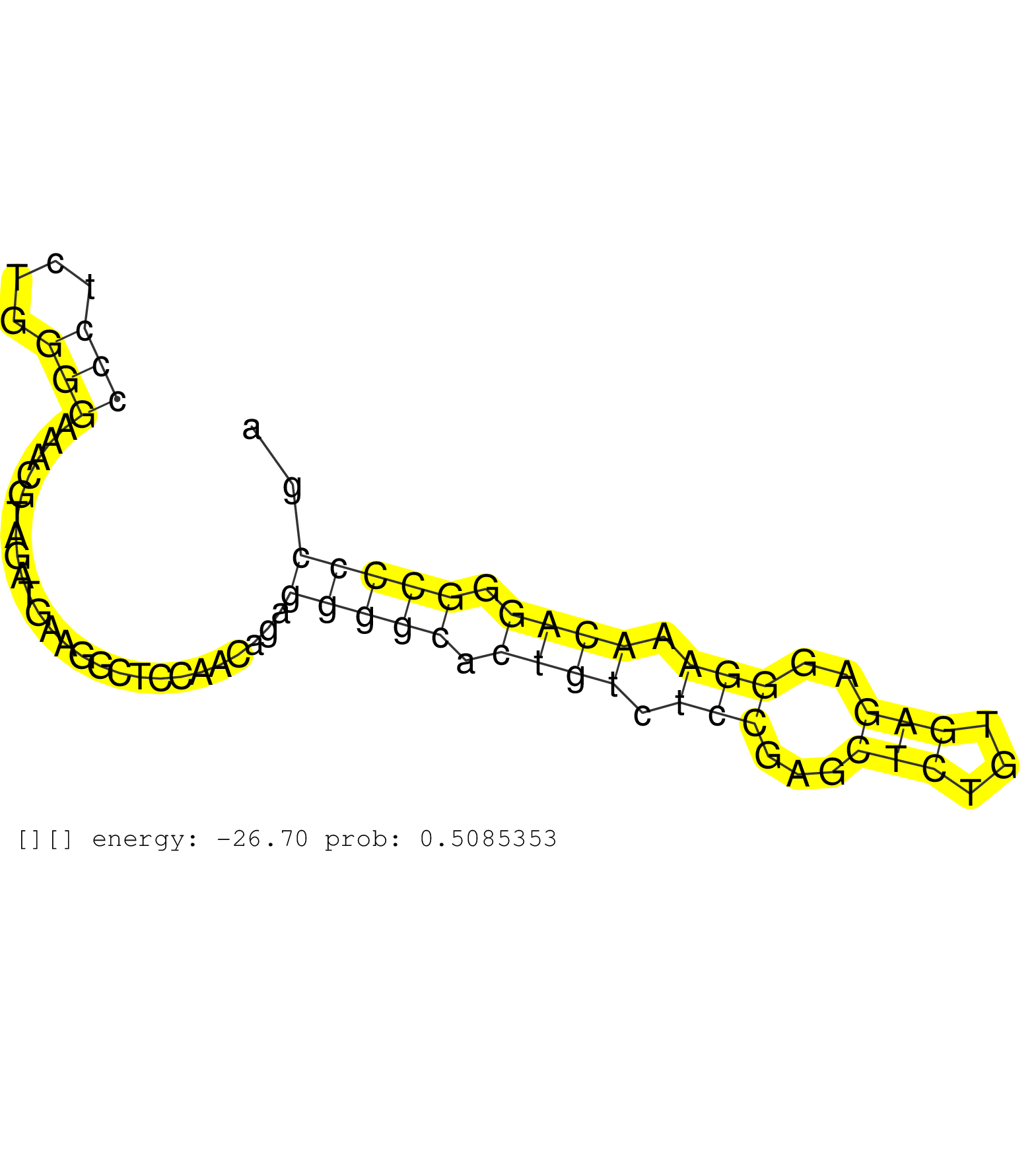
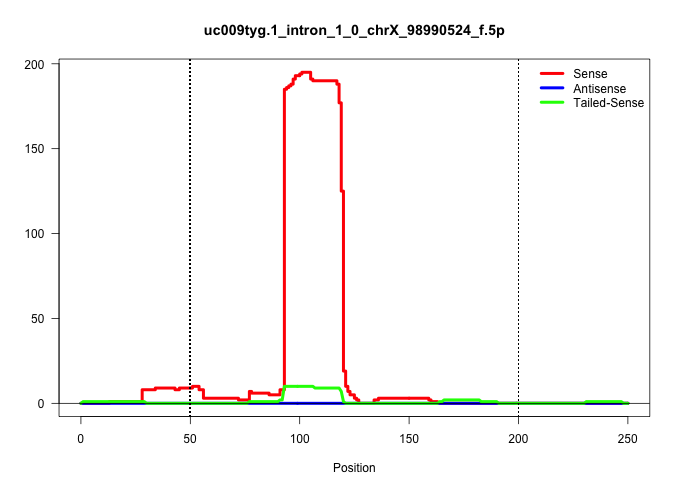
| Gene: BC100492 | ID: uc009tyg.1_intron_1_0_chrX_98990524_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| ATGACTGTCATGCGAGAGCTGACCAACCTACAGCTCCGAAAAGTCTGCAGGTACCCGTCCCCACGAACTTGTGCGGCTGAACTGGCTGCCCTCTGGGGAAACGTAGATGAAGGCTCCAACAGAGGGGCACTGTCTCCGAGCTCTGTGAGAGGGAAACAGGGCCCCGAAGGTGCACTGTACCCCAGAGGCCTGGGGCGAGGCAGAGCCTGGGGGACCCCAAGAAGACACACCATAGGTAGGATGGTGAGCA ........................................................................................(((....))).........................(((((.((((.(((...(((...)))..))).)))).)))))..................................................................................... ........................................................................................89............................................................................167................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAAC.................................................................................................................................. | 27 | 1 | 106.00 | 106.00 | 49.00 | 37.00 | 6.00 | 3.00 | 7.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAA................................................................................................................................... | 26 | 1 | 52.00 | 52.00 | 16.00 | 17.00 | 4.00 | 7.00 | 6.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAACA................................................................................................................................. | 28 | 1 | 9.00 | 9.00 | 1.00 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCA.................................................................................................................................... | 25 | 1 | 9.00 | 9.00 | 1.00 | 2.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TACAGCTCCGAAAAGTCTGCAGGTACCC.................................................................................................................................................................................................. | 28 | 1 | 5.00 | 5.00 | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACTGGCTGCCCTCTGGGGAAACGTA................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAAt.................................................................................................................................. | 27 | t | 3.00 | 52.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GAAACGTAGATGAAGGCTCCAACAGA............................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TACAGCTCCGAAAAGTCTGCAGGTAC.................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAAa.................................................................................................................................. | 27 | a | 2.00 | 52.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCTGGGGAAACGTAGATGAAGGCTCCA.................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CGAGCTCTGTGAGAGGGAAACAGGGCC....................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TCTGGGGAAACGTAGATGAAGGCTCC..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AAACGTAGATGAAGGCTCCAACAGAGGGG........................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCCGAGCTCTGTGAGAGGGAAACAGG.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................GGGAAACGTAGATGAAGGCTCCAACAG................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAACt................................................................................................................................. | 28 | t | 1.00 | 106.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGCAGGTACCCGTCCCCACGAACTTGT.................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................CGTAGATGAAGGCTCCAACAGAGGGG........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................TCCGAAAAGTCTGCAGGTACCCGTCCC............................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................ATAGGTAGGATGGTGcg.. | 17 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................ACGTAGATGAAGGCTCCAACAGAGGG............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACTGGCTGCCCTCTGGGGAAACGTAG................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GAAACGTAGATGAAGGCTCCAACAGAGG............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GGGGAAACGTAGATGAAGGCTCCAACAG................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AAACGTAGATGAAGGCTCCAACAGAGG............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................AAGGTGCACTGTAaggt................................................................... | 17 | aggt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TCCGAGCTCTGTGAGAGGGAAACAG........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCCAt................................................................................................................................... | 26 | t | 1.00 | 9.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCTGGGGAAACGTAGATGAAGGCTCCAt................................................................................................................................... | 28 | t | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GGAAACGTAGATGAAGGCTCCAACAG................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TGACTGTCATGCGAGAGCTGACCAACCTc............................................................................................................................................................................................................................ | 29 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................CACGAACTTGTGCGGCTGAACTGGC.................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TACCCGTCCCCACGAACTTGTGCGGCT............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACTGGCTGCCCTCTGGGGAAACGTAGc............................................................................................................................................... | 30 | c | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCaaaa.................................................................................................................................. | 27 | aaaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CGAAGGTGCACTGTACCCCAGAGGCCa........................................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................TGGGGAAACGTAGATGAAGGCTCC..................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............GAGAGCTGACCAACCTACAGCTCCGAAAAG............................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ATGACTGTCATGCGAGAGCTGACCAACCTACAGCTCCGAAAAGTCTGCAGGTACCCGTCCCCACGAACTTGTGCGGCTGAACTGGCTGCCCTCTGGGGAAACGTAGATGAAGGCTCCAACAGAGGGGCACTGTCTCCGAGCTCTGTGAGAGGGAAACAGGGCCCCGAAGGTGCACTGTACCCCAGAGGCCTGGGGCGAGGCAGAGCCTGGGGGACCCCAAGAAGACACACCATAGGTAGGATGGTGAGCA ........................................................................................(((....))).........................(((((.((((.(((...(((...)))..))).)))).)))))..................................................................................... ........................................................................................89............................................................................167................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|