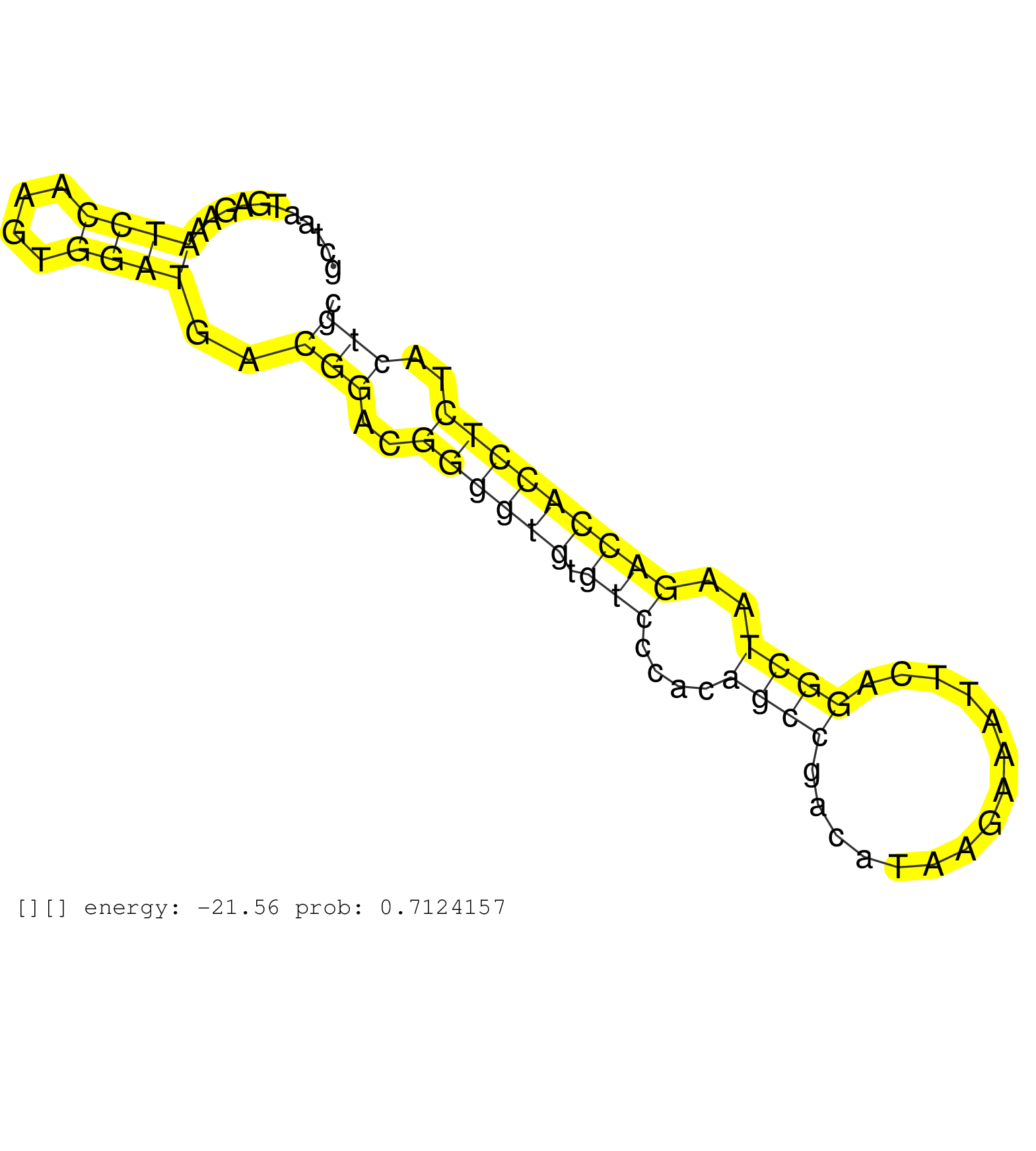
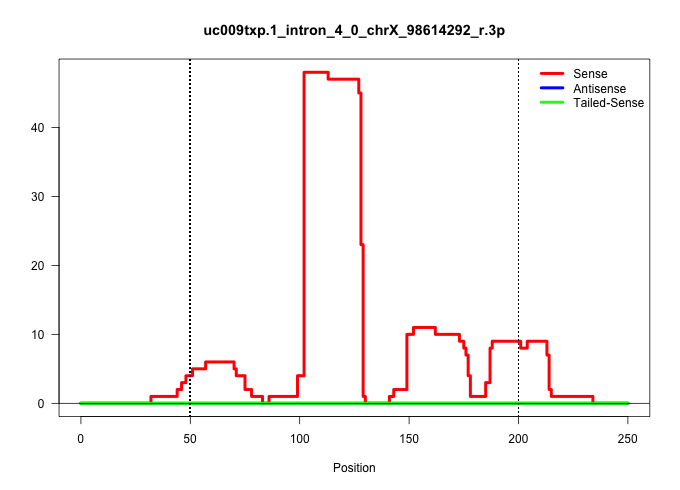
| Gene: Zmym3 | ID: uc009txp.1_intron_4_0_chrX_98614292_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| AGGGAACGCCACTCCCCACAGAGGGCGCTGTATTGGGGGGGGAGTGGACATTGAGCCTGAAGCCTGTGGAAATGCCTATCGGCTGTGGGGGGCGGCTGCTAATGAGAAATCCAAGTGGATGACGGACGGGGTGTGTCCCACAGCCGACATAAGAAATTCAGGCTAAGACCACCTCTACTGCCCCCTCTTCCGATCTGCAGCCCATACCCAGCCATGGACCCCAGTGATTTCCCCAGTCCATTTGACCCAT ............................................................................................................((((....))))..(((..((((((.(((....((((...............))))..)))))))))..)))...................................................................... .................................................................................................98.................................................................................181................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................TGAGAAATCCAAGTGGATGACGGACGG......................................................................................................................... | 27 | 1 | 22.00 | 22.00 | 6.00 | 8.00 | 2.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGAGAAATCCAAGTGGATGACGGACG.......................................................................................................................... | 26 | 1 | 21.00 | 21.00 | 6.00 | 5.00 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................CCGACATAAGAAATTCAGGCTAAGA.................................................................................. | 25 | 1 | 7.00 | 7.00 | - | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTCCGATCTGCAGCCCATACCCAGCCA.................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAAGAAATTCAGGCTAAGACCACCTCTA......................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TCTTCCGATCTGCAGCCCATACCCAGCC..................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAATGAGAAATCCAAGTGGATGACGGAC........................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAAGAAATTCAGGCTAAGACCACCTC........................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAAGAAATTCAGGCTAAGACCACCTCTAC........................................................................ | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCCGACATAAGAAATTCAGG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................TGAGAAATCCAAGTGGATGACGGACGGG........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGAAGCCTGTGGAAATGCCTATCGGC....................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TACCCAGCCATGGACCCCAGTGATTTCCCC................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................TCCGATCTGCAGCCCATACCCAGCCAT................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAATGAGAAATCCAAGTGGATGACGGACG.......................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TACTGCCCCCTCTTCCGATCTGCAGC................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................CATTGAGCCTGAAGCCTGTGGAAATGC............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................GACATTGAGCCTGAAGCCTGTGGAA................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................GTGGACATTGAGCCTGAAGCCTGTGG..................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................TGAGCCTGAAGCCTGTGGAAATGCCTA............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GAAATTCAGGCTAAGACCACCTCTAC........................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTGGGGGGGGAGTGGACA........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................GGGGGGCGGCTGCTAATGAGAAATCCA......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CCGACATAAGAAATTCAGGCTAAGACCACC............................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAAGAAATTCAGGCTAAGACCACCTCT.......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TTGAGCCTGAAGCCTGTGGAAATGC............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................TGGACATTGAGCCTGAAGCCTGTGGA.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGGAACGCCACTCCCCACAGAGGGCGCTGTATTGGGGGGGGAGTGGACATTGAGCCTGAAGCCTGTGGAAATGCCTATCGGCTGTGGGGGGCGGCTGCTAATGAGAAATCCAAGTGGATGACGGACGGGGTGTGTCCCACAGCCGACATAAGAAATTCAGGCTAAGACCACCTCTACTGCCCCCTCTTCCGATCTGCAGCCCATACCCAGCCATGGACCCCAGTGATTTCCCCAGTCCATTTGACCCAT ............................................................................................................((((....))))..(((..((((((.(((....((((...............))))..)))))))))..)))...................................................................... .................................................................................................98.................................................................................181................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|