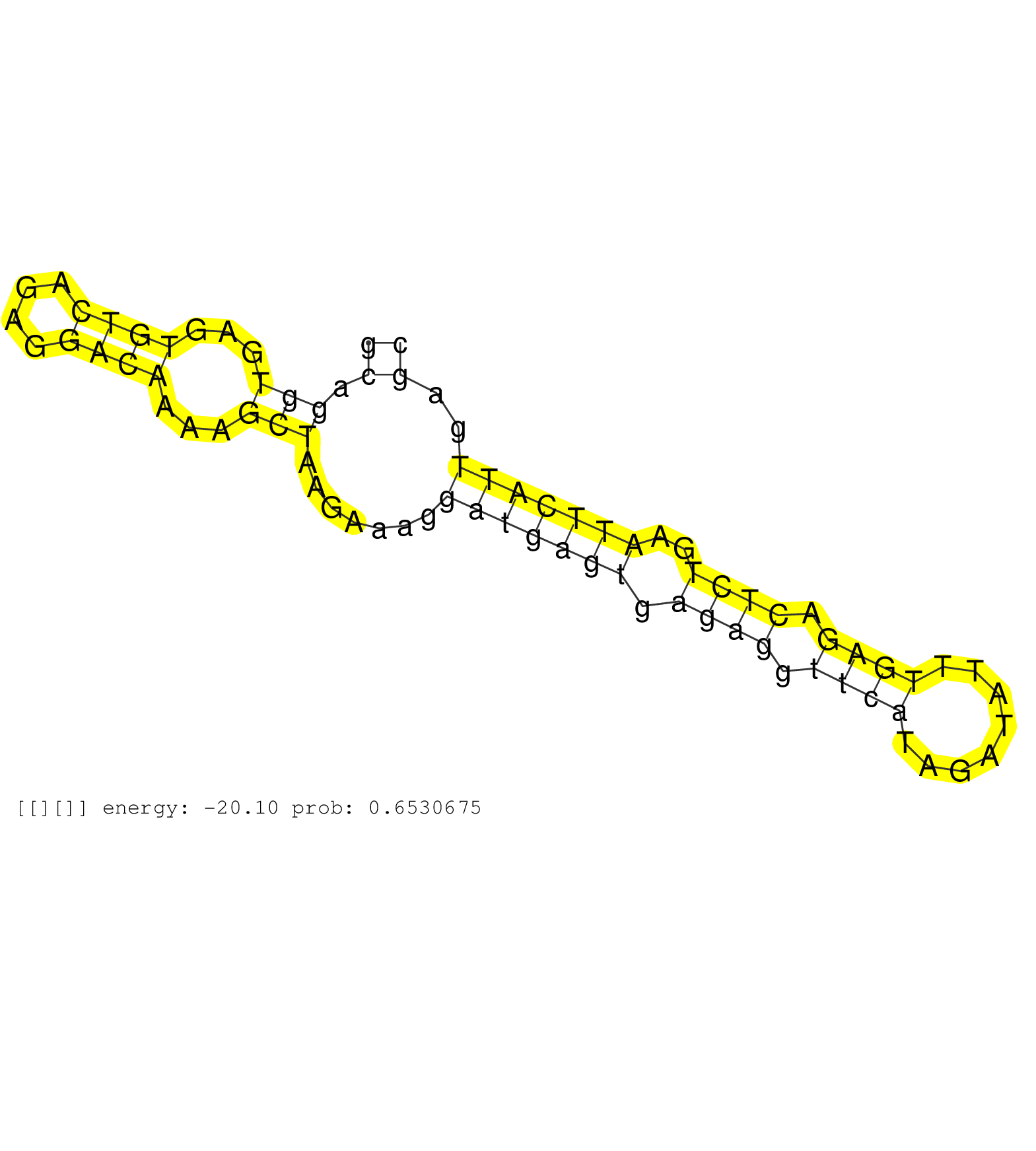
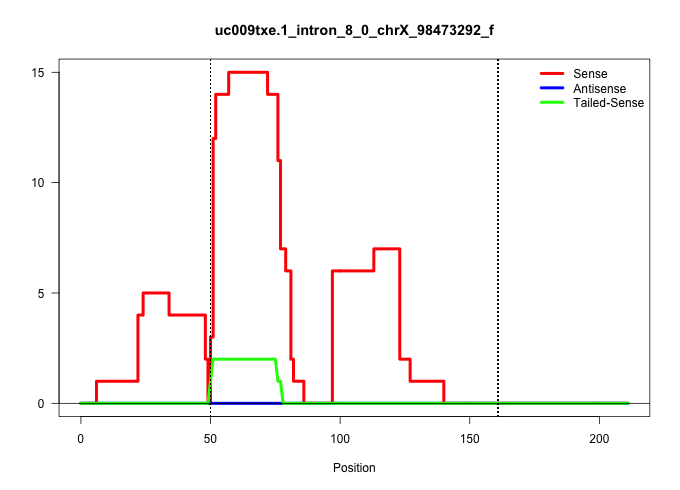
| Gene: Med12 | ID: uc009txe.1_intron_8_0_chrX_98473292_f | SPECIES: mm9 |
 |
 |
|
(6) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| AGCAGTTGAGGTTCGCTGGTCTTTTGATAAGTGCCAGGAAGCTACTGCAGGTGAGTGTCAGAGGACAAAAGCTAAGAAAGGATGAGTGAGAGGTTCATAGATATTTGAGACTCTGAATTCATTGAGCCTCTAGTTGAGTCACCTTTACTGTTTGTCCTCAGGTTTCACCATTGGACGGGTGCTCCATACTTTAGAAGTGCTGGATAGCCAT ..............................................((.(((...((((....))))...))).......(((((((.((((.((((........)))).))))..)))))))..)).................................................................................... ..............................................47..............................................................................127.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................TAGATATTTGAGACTCTGAATTCATT........................................................................................ | 26 | 1 | 5.00 | 5.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAGA...................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAGAAAGG.................................................................................................................................. | 30 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTTGATAAGTGCCAGGAAGCTACTGC................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTCAGAGGACAAAAGCTAAG....................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................GAGTGTCAGAGGACAAAAGCTAAGA...................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TCAGAGGACAAAAGCTAAGAAAGGATGAG............................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAG....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGATAAGTGCCAGGAAGCTACTGCA.................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAGAAAGGA................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGAGTGTCAGAGGACAAAAGC........................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAt....................................................................................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................TGAGTGTCAGAGGACAAAAGCTAAGAAA.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ......TGAGGTTCGCTGGTCTTTTGATAAGTGC................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGAATTCATTGAGCCTCTAGTTGAGTC....................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................TTTGATAAGTGCCAGGAAGCTACTGCA.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................TAGATATTTGAGACTCTGAATTCATTGAGC.................................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTCAGAGGACAAAAGCTAAGAt..................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GAGTGTCAGAGGACAAAAGCTAAGAAAGG.................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCAGTTGAGGTTCGCTGGTCTTTTGATAAGTGCCAGGAAGCTACTGCAGGTGAGTGTCAGAGGACAAAAGCTAAGAAAGGATGAGTGAGAGGTTCATAGATATTTGAGACTCTGAATTCATTGAGCCTCTAGTTGAGTCACCTTTACTGTTTGTCCTCAGGTTTCACCATTGGACGGGTGCTCCATACTTTAGAAGTGCTGGATAGCCAT ..............................................((.(((...((((....))))...))).......(((((((.((((.((((........)))).))))..)))))))..)).................................................................................... ..............................................47..............................................................................127.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|