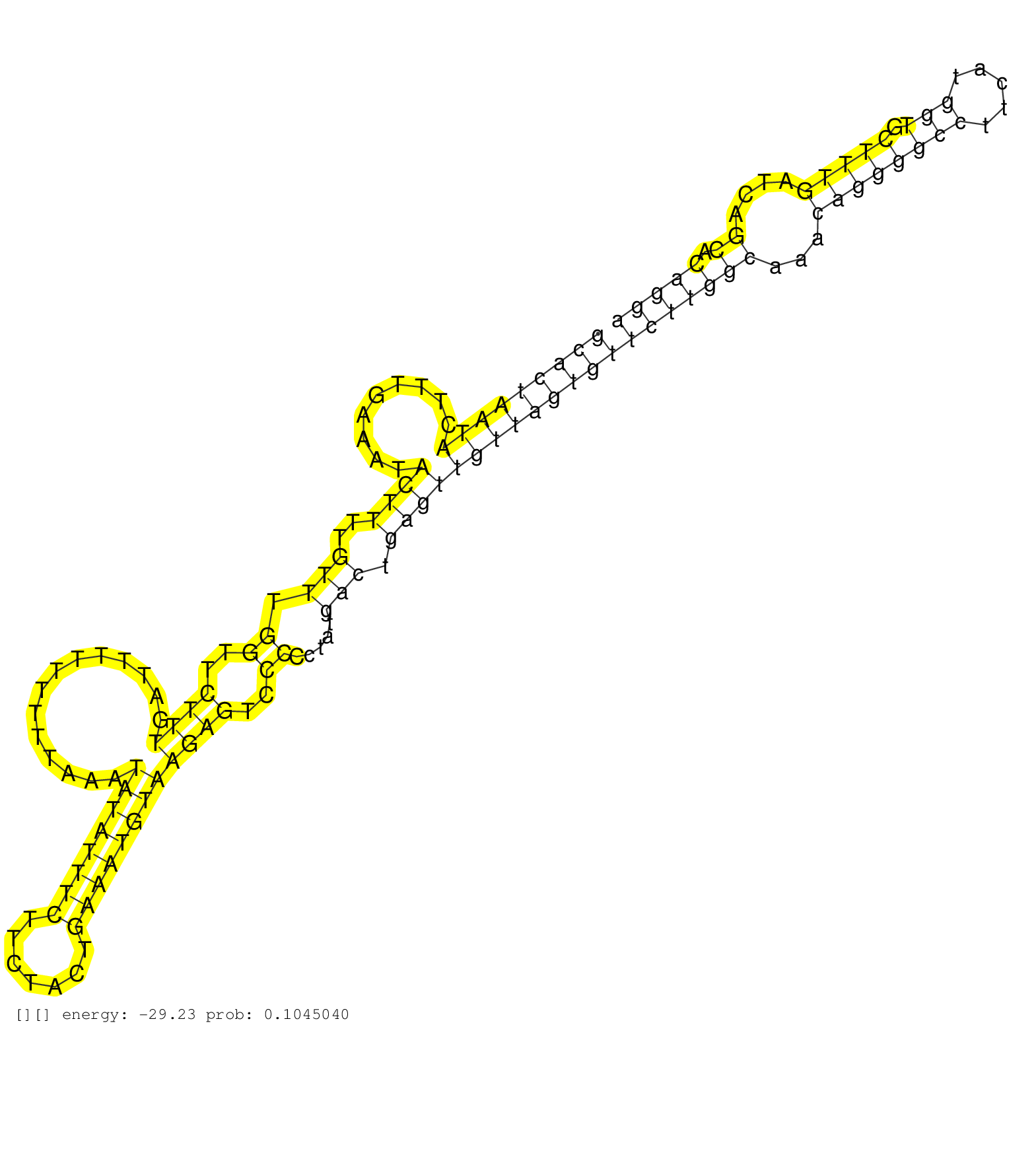

| Gene: Yipf6 | ID: uc009tvc.1_intron_2_0_chrX_96135241_f | SPECIES: mm9 |
 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| CTCAAGAACATGACAGCTCCACACTGAATGAATCCATTCGTCGGACCATTGTAAGTTAGACGAGTTGAGAGACCACTTCAGTGTTATGGGAGCGAATAGGCACTAATACTTTGAAATACTTTTGTTTGGTTCTTTGATTTTTTTTTAAATATATTTCTTCTACTGAAATGTAAGAGTCCCCCTATGACTGAGTTGTTAGTGTTCTTGGCAAACAGGGGCCTTCATGGTGCTTTGATCAGCACAGGACATGGAGTTATTTTTGTTTTTCAGATGCGAGATCTAAAAGCTGTGGGAAGGAAATTTATGCATGTTTTATATCC ...................................................................................................(((((((((.........((((..(((.((..((((..............((((((((.......))))))))))))..)).....))).)))))))))))))(((((((...((((((((.....))).)))))....)).))))).......................................................................... ...................................................................................................100................................................................................................................................................246........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................AATACTTTGAAATACTTTTGTTTGGTTCTTTGATTTTTTTTTAAATATATTT.................................................................................................................................................................... | 52 | 1 | 43.00 | 43.00 | 43.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TGCGAGATCTAAAAGCTGTGGGAAGGA...................... | 27 | 1 | 6.00 | 6.00 | - | 2.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - |
| .TCAAGAACATGACAGCTCCA........................................................................................................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................TAAAAGCTGTGGGAAGGAAATTTATGt............. | 27 | t | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................CGAGATCTAAAAGCTGTGGGAAGGAAATa.................. | 29 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..CAAGAACATGACAGCTCCACACTGAA.................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................TAAAAGCTGTGGGAAGGAAATTTttt.............. | 26 | ttt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..CAAGAACATGACAGCTCCACACTGAAT................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................TAAAAGCTGTGGGAAGGAAATTTATG.............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................ATGAATCCATTCGattc.................................................................................................................................................................................................................................................................................... | 17 | attc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................................................................................TGCGAGATCTAAAAGCTGTGGGAAGGAA..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................................................................................TAAAAGCTGTGGGAAGGAAATTTATGC............. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................TGCGAGATCTAAAAGCTGTGGGAAGGAAA.................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................................................................................TAAAAGCTGTGGGAAGGAAATTTATGCAT........... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CTCAAGAACATGACAGCTCCACACTGAATGAATCCATTCGTCGGACCATTGTAAGTTAGACGAGTTGAGAGACCACTTCAGTGTTATGGGAGCGAATAGGCACTAATACTTTGAAATACTTTTGTTTGGTTCTTTGATTTTTTTTTAAATATATTTCTTCTACTGAAATGTAAGAGTCCCCCTATGACTGAGTTGTTAGTGTTCTTGGCAAACAGGGGCCTTCATGGTGCTTTGATCAGCACAGGACATGGAGTTATTTTTGTTTTTCAGATGCGAGATCTAAAAGCTGTGGGAAGGAAATTTATGCATGTTTTATATCC ...................................................................................................(((((((((.........((((..(((.((..((((..............((((((((.......))))))))))))..)).....))).)))))))))))))(((((((...((((((((.....))).)))))....)).))))).......................................................................... ...................................................................................................100................................................................................................................................................246........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|