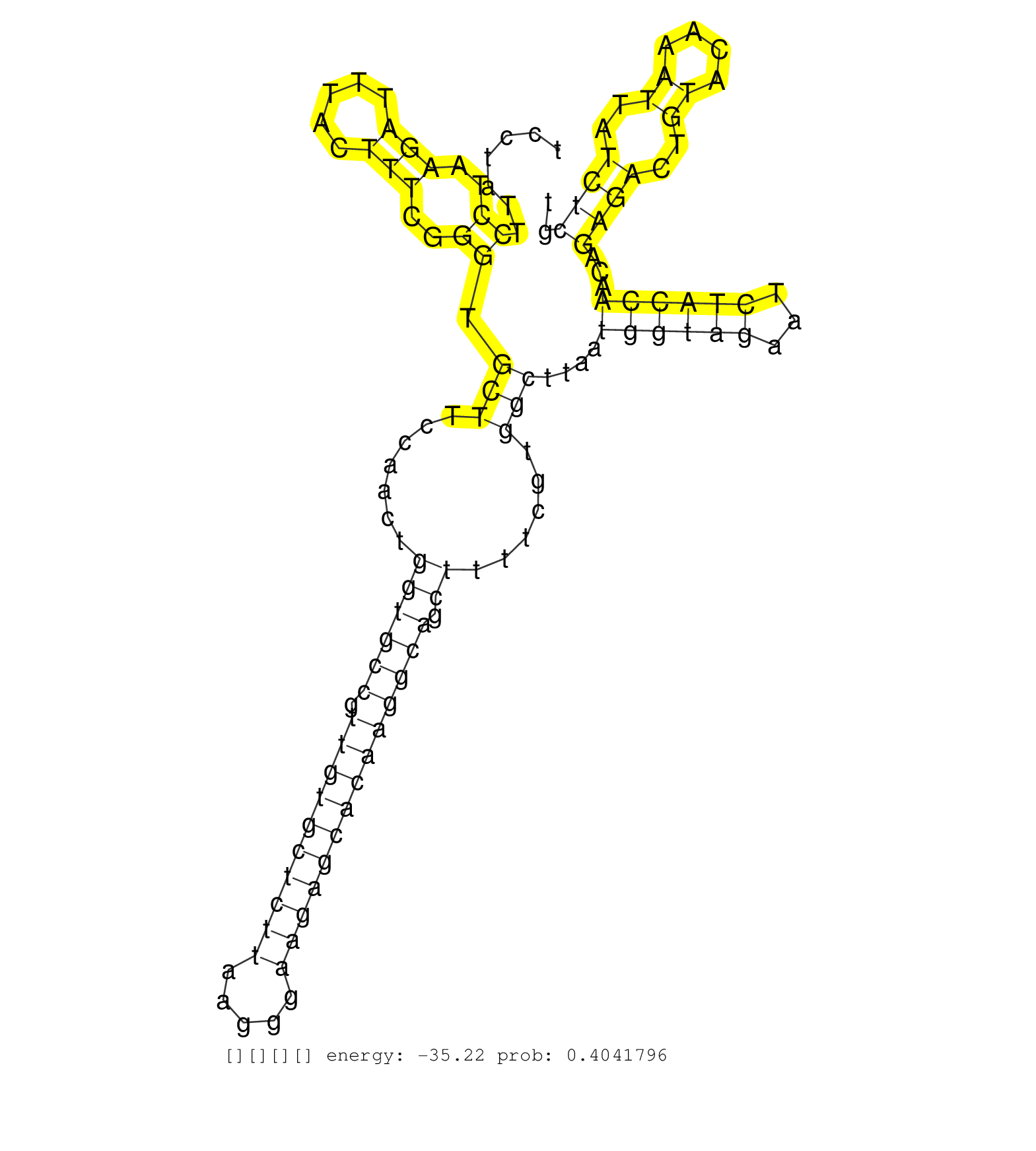
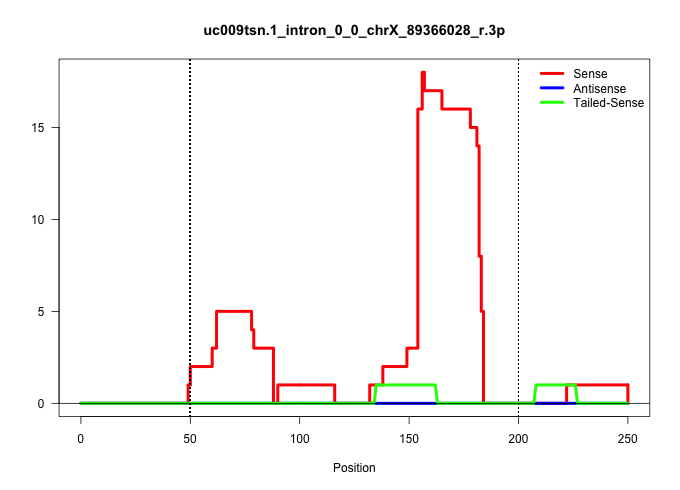
| Gene: Mageb18 | ID: uc009tsn.1_intron_0_0_chrX_89366028_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(3) PIWI.mut |
(14) TESTES |
| CATTCTTACCCCACAGACCAGAGGCTTTCCCAAAGGGACTCTCTCCATTTTATCCAGTCCTATTCCTAAGATTTACTTTCGGGTGCTTCCAACTGGTGCCGTTGTGCTCTTAAGGGAAGAGCACAAGGCAGCTTTTCGTGGCTTAATGGTAGAATCTACCAACAGAGACTGTACAAATTATCTCGTCCTCTGTTTTCCAGGGTGACCTCTTCCTCAGCCTGTTTGCAGTACTACTTGTGATCCCCACTAC ................................................................((..(((.....)))..)).(((.......((((((.((((((((((.....)))))))))))))).))......)))....((((((...))))))...((((..((....))..)))).................................................................. .........................................................58..............................................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................TCTACCAACAGAGACTGTACAAATTATC.................................................................... | 28 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................TCTACCAACAGAGACTGTACAAATTATCTC.................................................................. | 30 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................................TCTACCAACAGAGACTGTACAAATTATCT................................................................... | 29 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................TATTCCTAAGATTTACTTTCGGGTGCTT.................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................TTCCTAAGATTTACTTTCGGGTGCTT.................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................TACCAACAGAGACTGTACAAATTATCTC.................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TCGTGGCTTAATGGTAGAATCTACCAAa....................................................................................... | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTTCGTGGCTTAATGGTAGAATCT............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AACTGGTGCCGTTGTGCTCTTAAGGG...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCTACCAACAGAGACTGTACAAATTAT..................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGGCTTAATGGTAGAATCTACCAACAG..................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTGCAGTACTACTTGTGATCCCCACTAC | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TATCCAGTCCTATTCCTAAGATTTACTT............................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TCGGGTGCTTCCAACTGGTGCCGTTGT................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAGAATCTACCAACAGAGACTGTACAAAT........................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................CAAATTATCTCGTCCTCTGTTTTtcac.................................................. | 27 | tcac | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TTATCCAGTCCTATTCCTAAGATTTACTTT........................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................................................CTTCCTCAGCCTGTTgggg....................... | 19 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CATTCTTACCCCACAGACCAGAGGCTTTCCCAAAGGGACTCTCTCCATTTTATCCAGTCCTATTCCTAAGATTTACTTTCGGGTGCTTCCAACTGGTGCCGTTGTGCTCTTAAGGGAAGAGCACAAGGCAGCTTTTCGTGGCTTAATGGTAGAATCTACCAACAGAGACTGTACAAATTATCTCGTCCTCTGTTTTCCAGGGTGACCTCTTCCTCAGCCTGTTTGCAGTACTACTTGTGATCCCCACTAC ................................................................((..(((.....)))..)).(((.......((((((.((((((((((.....)))))))))))))).))......)))....((((((...))))))...((((..((....))..)))).................................................................. .........................................................58..............................................................................................................................186.............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|