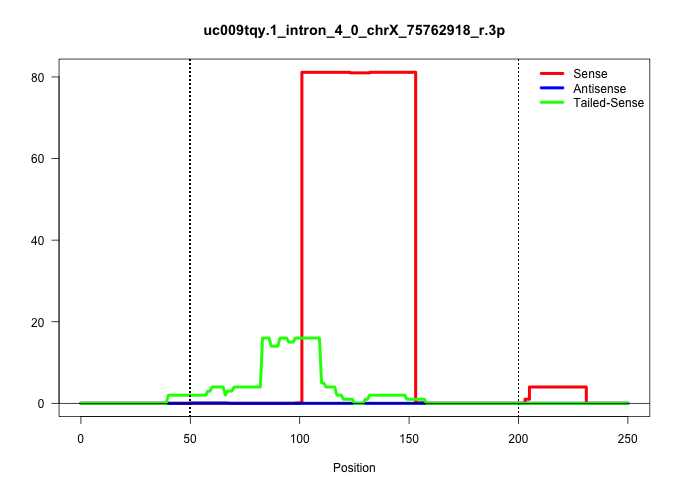
| Gene: Prrg1 | ID: uc009tqy.1_intron_4_0_chrX_75762918_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(3) PIWI.mut |
(15) TESTES |
| TACACCTCGCGTCCATCGTGAGAACTGTGCAGTTGCACCATGATGCAAGATCAGACCATATGACAAGTGATGATTCTGGCCAATGGACTGCTGTTACGTGGACTGGGCTGATGGGGCGTGGTAGAGAGGTTATATAAGGGATTGTGATTGGGGGTGGAAAAAAGAGAAGAGAGATTTTTTTCCTGCATCCATGTTGAAAGGTTCCTGAAAAAACTGCTTTGAGAAGGACGCTGTGTCGTAGCTCTTTTCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................ACTGGGCTGATGGGGCGTGGTAGAGAGGTTATATAAGGGATTGTGATTGGGG................................................................................................. | 52 | 1 | 81.00 | 81.00 | 81.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGGACTGCTGTTACGTGGACTGGGCat............................................................................................................................................ | 27 | at | 9.00 | 0.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAAAAAACTGCTTTGAGAAGGACGC................... | 26 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................TGATGCAAGATCAGACCATATGACgt........................................................................................................................................................................................ | 26 | gt | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TATGACAAGTGATGATTCTGGCCAATGGt................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGACAAGTGATGATTCTGGCCAATGGc................................................................................................................................................................... | 27 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................TGGACTGCTGTTACGTGGACTGGGCTa............................................................................................................................................ | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................TGATGATTCTGGCCAATGGACTGCTGa............................................................................................................................................................ | 27 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TATATAAGGGATTGTagct..................................................................................................... | 19 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................TGTTACGTGGACTGGGCTGATGGGaa..................................................................................................................................... | 26 | aa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTTACGTGGACTGGGCTGATGcggc..................................................................................................................................... | 26 | cggc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TATAAGGGATTGTGATTGGGGGgaga............................................................................................ | 26 | gaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................TGGACTGCTGTTACGTGGACTGGcaag............................................................................................................................................ | 27 | caag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................TGGACTGGGCTGATGGGGCGTGGattt............................................................................................................................. | 27 | attt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................TGATTCTGGCCAATGGACTGCTGct........................................................................................................................................................... | 25 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................CCTGAAAAAACTGCTTTGAGAAGGACGC................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TACGTGGACTGGGCTGATGGGGCGTt.................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................TGGACTGCTGTTACGTGGACTGGGCTGAa.......................................................................................................................................... | 29 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TATAAGGGATTGTGATTGGGGGTGG............................................................................................. | 25 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGGACTGGGCTGATGGGGCGTGGTA............................................................................................................................... | 25 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 | - |
| ....................................................................................................GACTGGGCTGATGGGGCGTGGTA............................................................................................................................... | 23 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| TACACCTCGCGTCCATCGTGAGAACTGTGCAGTTGCACCATGATGCAAGATCAGACCATATGACAAGTGATGATTCTGGCCAATGGACTGCTGTTACGTGGACTGGGCTGATGGGGCGTGGTAGAGAGGTTATATAAGGGATTGTGATTGGGGGTGGAAAAAAGAGAAGAGAGATTTTTTTCCTGCATCCATGTTGAAAGGTTCCTGAAAAAACTGCTTTGAGAAGGACGCTGTGTCGTAGCTCTTTTCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TCAGACCATATGACAAGT...................................................................................................................................................................................... | 18 | 14 | 0.07 | 0.07 | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - |