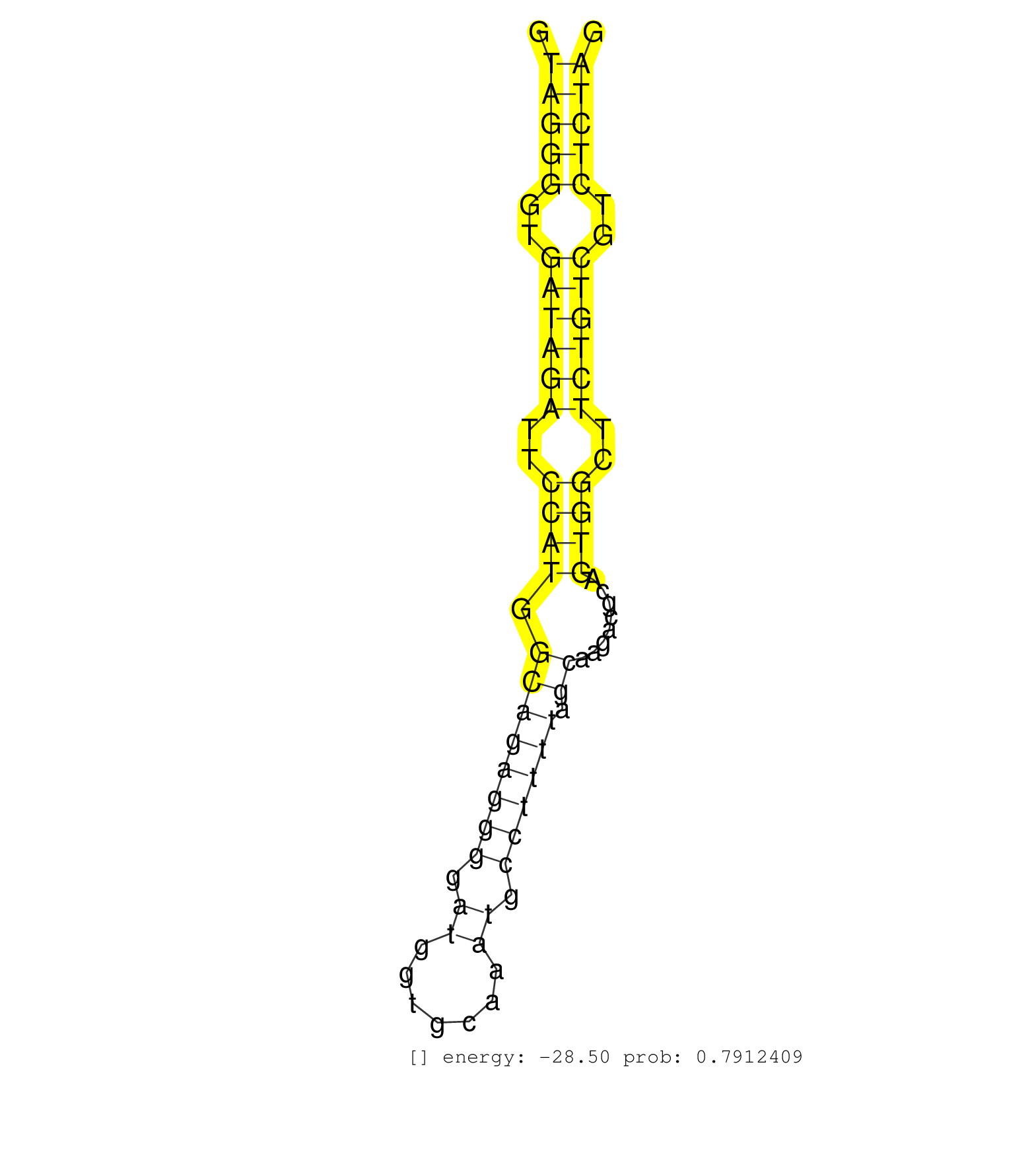
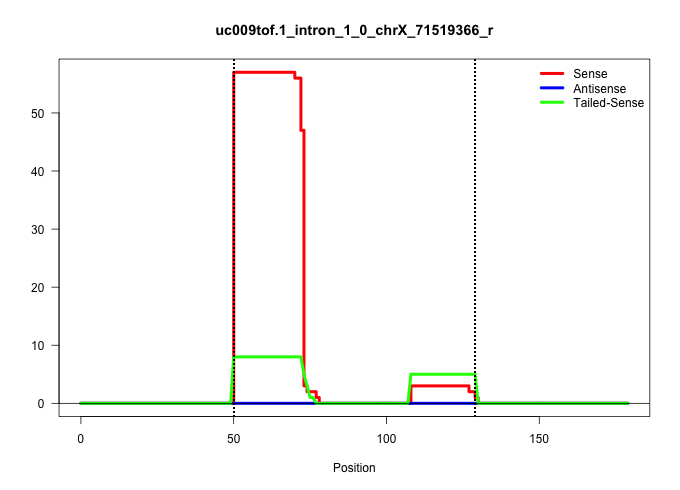
| Gene: Dnase1l1 | ID: uc009tof.1_intron_1_0_chrX_71519366_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) OVARY |
(2) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| ATGCCCTCTATGATGTGTTTCTGGATGTCTACCAGCGCTGGCAGAATGAGGTAGGGGTGATAGATTCCATGGCAGAGGGGATGGTGCAAATGCCTTTTAGCAAGACGCAGTGGCTTCTGTCGTCTCTAGAATGTGATTCTGCTTGGAGACTTCAATGCAGACTGTGCATCGCTGACAAA ...................................................(((((..((((((..((((.((((((((.((.......)).)))))).))........))))..))))))..)))))................................................... ..................................................51............................................................................129................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGGGTGATAGATTCCATGGC.......................................................................................................... | 23 | 1 | 62.00 | 62.00 | 20.00 | 11.00 | 9.00 | 5.00 | 7.00 | 1.00 | - | 3.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 |
| ..................................................GTAGGGGTGATAGATTCCATGG........................................................................................................... | 22 | 1 | 16.00 | 16.00 | 3.00 | - | 1.00 | 4.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGTGGCTTCTGTCGTCTCTAGt................................................. | 22 | t | 7.00 | 2.00 | - | - | - | - | - | - | 3.00 | - | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................AGTGGCTTCTGTCGTCTCTAGA................................................. | 22 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCAt........................................................................................................ | 25 | t | 4.00 | 2.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGa.......................................................................................................... | 23 | a | 3.00 | 16.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCA......................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGt.......................................................................................................... | 23 | t | 2.00 | 16.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGTGGCTTCTGTCGTCTCTAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGaa......................................................................................................... | 24 | aa | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCAGAG...................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCAttt...................................................................................................... | 27 | ttt | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCAGAGG..................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCAGt....................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCt......................................................................................................... | 24 | t | 1.00 | 62.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCAT............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGGGTGATAGATTCCATGGCtt........................................................................................................ | 25 | tt | 1.00 | 62.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGCGCTGGCAGAATGAG................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGTGGCTTCTGTCGTCTCT.................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATGCCCTCTATGATGTGTTTCTGGATGTCTACCAGCGCTGGCAGAATGAGGTAGGGGTGATAGATTCCATGGCAGAGGGGATGGTGCAAATGCCTTTTAGCAAGACGCAGTGGCTTCTGTCGTCTCTAGAATGTGATTCTGCTTGGAGACTTCAATGCAGACTGTGCATCGCTGACAAA ...................................................(((((..((((((..((((.((((((((.((.......)).)))))).))........))))..))))))..)))))................................................... ..................................................51............................................................................129................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|