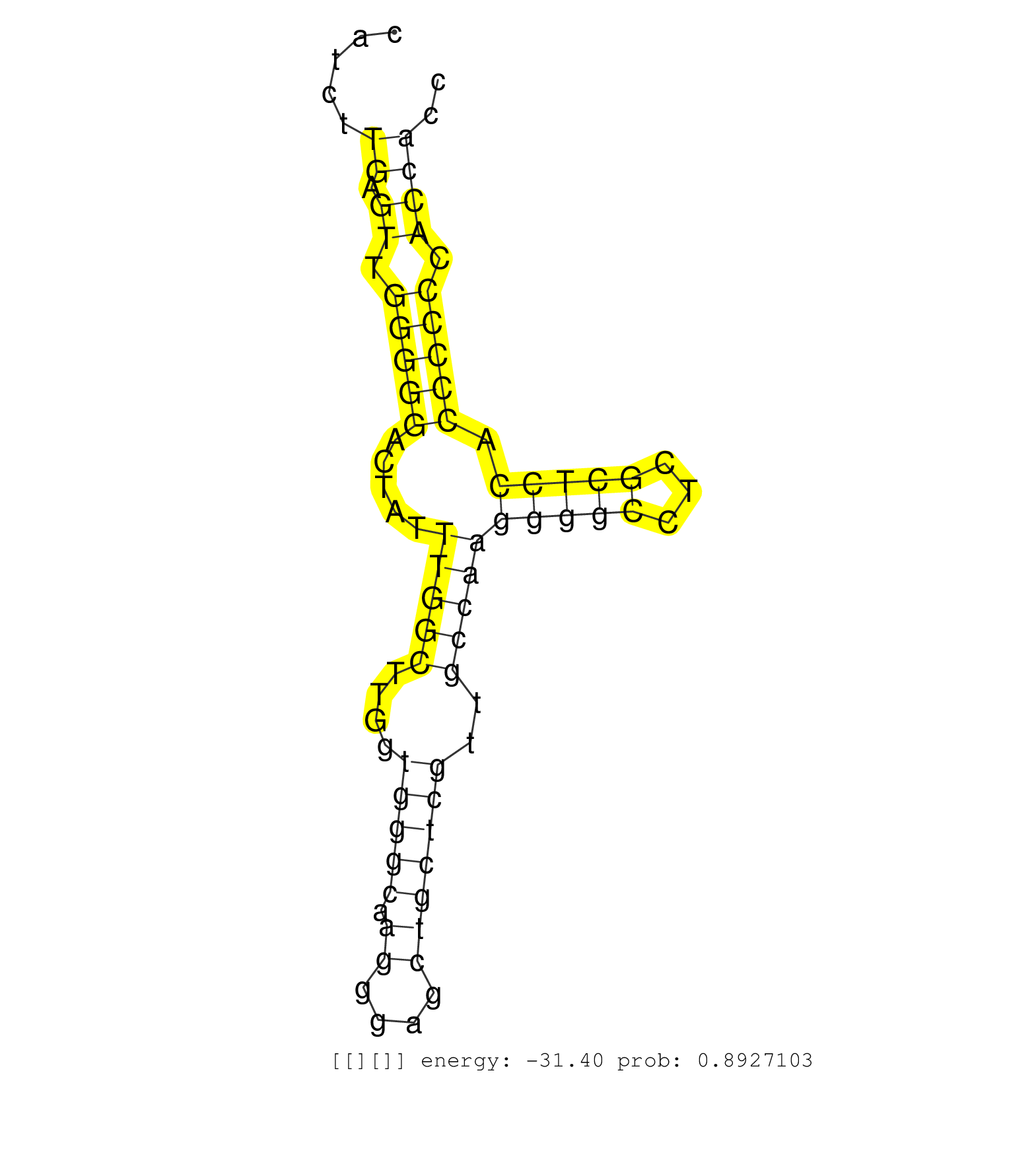

| Gene: Pnck | ID: uc009tme.1_intron_10_0_chrX_70904589_r.3p | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(2) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(22) TESTES |
| ATACCTCGGTGGGAAGCCTGCATGCAGTGCTCTGCCTCAGTGGGAGTGTGTGTGTGTGGCCCTGGGCGGGAGGAGTAAGGTGTACTTGGGCTTGGGAGTGGGTGACTCCATCTTGAGTTGGGGGACTATTTGGCTTGGTGGGCAAGGGAGCTGCTCGTTGCCAAGGGGCCTCGCTCCACCCCCCACCACCCCACCCCCAGACATGCTGCTGCTCAAGAAACAGACGGAGGACATCAGCAGTGTCTATGAG .................................................................................................................((.((.(((((.....(((((....(((((.((....)))))))..)))))(((((...))))).))))).)))).............................................................. ............................................................................................................109..............................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037896(GSM510432) ovary_rep1. (ovary) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................CCTCGCTCCACCCCCCca................................................................ | 18 | ca | 15.00 | 0.00 | 3.00 | - | 5.00 | - | 1.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCca................................................................ | 22 | ca | 13.00 | 8.00 | 3.00 | 1.00 | 1.00 | 3.00 | - | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GCCTCGCTCCACCCCCCca................................................................ | 19 | ca | 11.00 | 0.00 | 3.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 3.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCC.................................................................. | 20 | 1 | 8.00 | 8.00 | - | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCC................................................................... | 19 | 1 | 7.00 | 7.00 | - | 5.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCc................................................................. | 21 | c | 6.00 | 8.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 |
| ......................................................................................................................................................................GGCCTCGCTCCACCCCCCca................................................................ | 20 | ca | 6.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCCCca................................................................ | 21 | ca | 6.00 | 0.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCtcca................................................................ | 22 | tcca | 3.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCC................................................................... | 18 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGAGTTGGGGGACTATTTGGCTTG................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GGCCTCGCTCCACCCCCCc................................................................. | 19 | c | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCccc............................................................... | 23 | ccc | 2.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCC.................................................................... | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GCCTCGCTCCACCCCtccc................................................................ | 19 | tccc | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GGCCTCGCTCCACCCCCCcca............................................................... | 21 | cca | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCcat............................................................... | 23 | cat | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCcg................................................................ | 22 | cg | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CCTCGCTCCACCCCCCcca............................................................... | 19 | cca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACaat.................................................................... | 18 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................CCTCGCTCCACCCCCCcat............................................................... | 19 | cat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CCTCGCTCCACCCCCtcca............................................................... | 19 | tcca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CTCGCTCCACCCCCCcca............................................................... | 18 | cca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGCTCAAGAAACAGACGGAGGACATCAG............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................GCCTCGCTCCACCCCCCcatc.............................................................. | 21 | catc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCA................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................CGCTCCACCCCCCccat.............................................................. | 17 | ccat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GGGGCCTCGCTCCACCCCCCcca............................................................... | 23 | cca | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCCCcca............................................................... | 22 | cca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCCCta................................................................ | 21 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCCCcatc.............................................................. | 23 | catc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GCCTCGCTCCACCCCCCc................................................................. | 18 | c | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TCGCTCCACCCCCCccca.............................................................. | 18 | ccca | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GGGCCTCGCTCCACCCCCCcat............................................................... | 22 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CCTCGCTCCACCCCCCta................................................................ | 18 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CTCGCTCCACCCCCCccca.............................................................. | 19 | ccca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................GGCCTCGCTCCACCCCCCA................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................GCCTCGCTCCACCCCCCcca............................................................... | 20 | cca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TACCTCGGTGGGAAGCCTGCATGCAGT.............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ATACCTCGGTGGGAAGCCTGCATGCAGTGCTCTGCCTCAGTGGGAGTGTGTGTGTGTGGCCCTGGGCGGGAGGAGTAAGGTGTACTTGGGCTTGGGAGTGGGTGACTCCATCTTGAGTTGGGGGACTATTTGGCTTGGTGGGCAAGGGAGCTGCTCGTTGCCAAGGGGCCTCGCTCCACCCCCCACCACCCCACCCCCAGACATGCTGCTGCTCAAGAAACAGACGGAGGACATCAGCAGTGTCTATGAG .................................................................................................................((.((.(((((.....(((((....(((((.((....)))))))..)))))(((((...))))).))))).)))).............................................................. ............................................................................................................109..............................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR037896(GSM510432) ovary_rep1. (ovary) |
|---|