
| Gene: Utp14a | ID: uc009tcb.1_intron_9_0_chrX_45627154_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
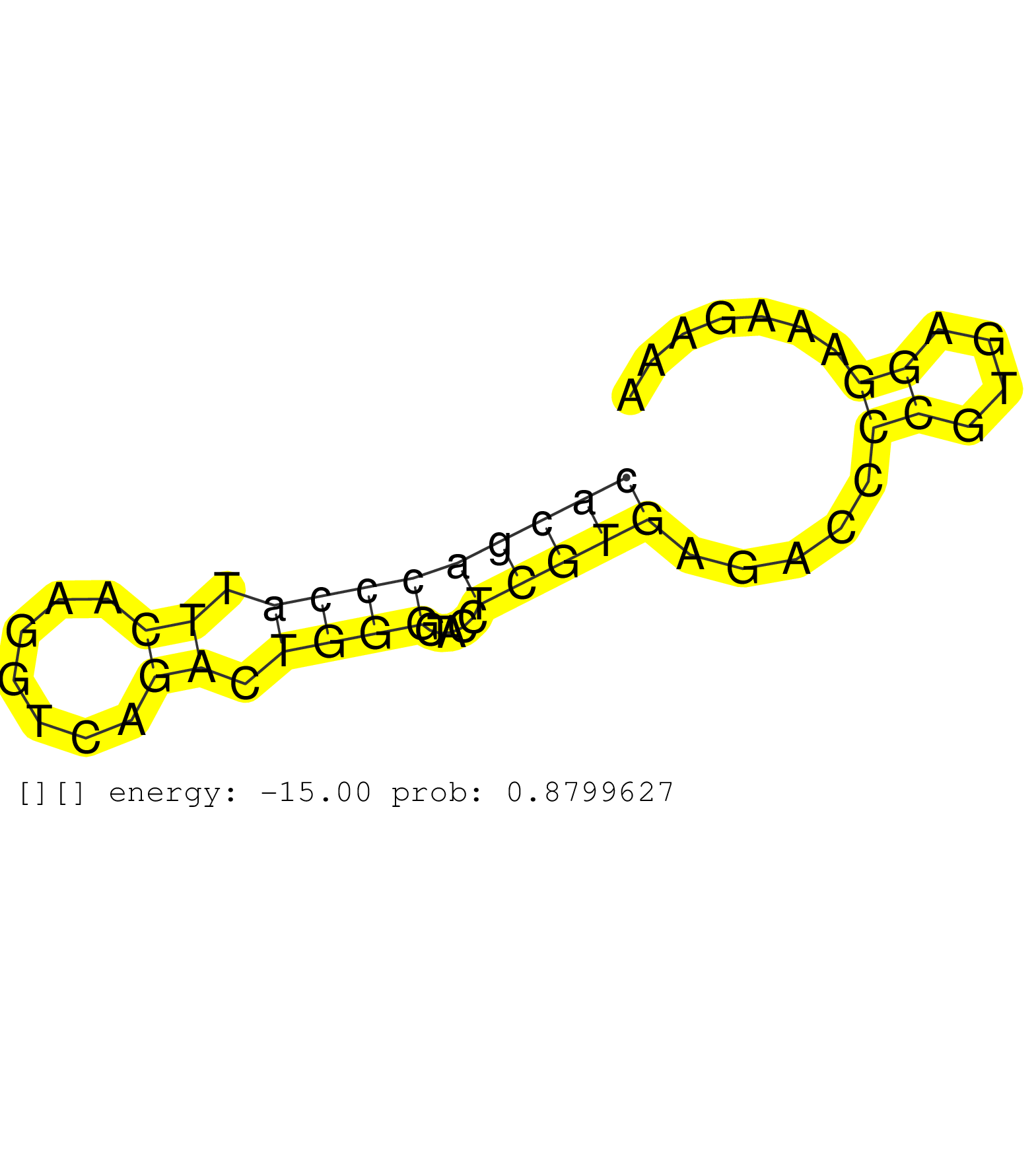
| CAGAAATCTTGGGCTGCCTACCTGTGAAGCCAGCACTAGAAAGGCTGAGGCACGACCCATTCAAGGTCAGACTGGGCTACCTCGTGAGACCCCGTGAGGAAAGAAAGGCCGCTTTAAGAAGTGAGGGGCAAGCAAGGTTCTTGGTCATTTCTGTGGTCAGTAATGACAAAGCTCCTGCACTGGGGCACTCCTTCCTCTAGGCCCGCCAAGCTATGCAAGAGCAGTTGGCTAAGAACAAGGAACTGACCCA ..................................................(((((((((.((.......)).)))).....))))).....((....))....................................................................................................................................................... ..................................................51.....................................................106.............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAG........... | 26 | 1 | 20.00 | 20.00 | 8.00 | 3.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAGG.......... | 27 | 1 | 13.00 | 13.00 | - | 3.00 | - | 2.00 | 1.00 | 1.00 | 2.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................TAGAAAGGCTGAGGCACGACCCATTC............................................................................................................................................................................................ | 26 | 1 | 6.00 | 6.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................TTCAAGGTCAGACTGGGCTACCTCGTG.................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTCAAGGTCAGACTGGGCTACCTCGT..................................................................................................................................................................... | 26 | 1 | 6.00 | 6.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAGGA......... | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGAGACCCCGTGAGGAAAGAAAGGCCG........................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAGGt......... | 28 | t | 3.00 | 13.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGGCCCGCCAAGCTATGCAAGAGCAGTT........................ | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TTCAAGGTCAGACTGGGCTACCTCGTa.................................................................................................................................................................... | 27 | a | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - |
| ..GAAATCTTGGGCTGCCTACCTGTGA............................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGGGCTGCCTACCTGTGAAGCCAGCACT..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAGGc......... | 28 | c | 1.00 | 13.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TTAAGAAGTGAGGGGCAAGCAAGGa................................................................................................................ | 25 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TAAGAAGTGAGGGGCAAGCAAGGTT............................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AAGAAGTGAGGGGCAAGCAAGGTTC.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................AGGAAAGAAAGGCCGCTTTAAGAAGTG............................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................TTTAAGAAGTGAGGatca........................................................................................................................ | 18 | atca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................GCAAGAGCAGTTGGCTAAGAACAAGa.......... | 26 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TTGGCTAAGAACAAGagga....... | 19 | agga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................TACCTGTGAAGCCAGCACTAGAAAGGCTGAGGC....................................................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TCTTGGGCTGCCTACCTGTGAAGCCt.......................................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................TTTAAGAAGTGAGGGGCAAGCAAGGT................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TACCTGTGAAGCCAGCACTAGAAAGG.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................TACCTGTGAAGCCAGCACTAGAAAGGCTG........................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................GAAAGGCTGAGGCACGACCCATTCAAGG........................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................TTCTTGGTCATTTCTGggt.............................................................................................. | 19 | ggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........TGGGCTGCCTACCTGTGAAGCCAGCAC...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................CCATTCAAGGTCAGACTGGGCTACCTC....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGCAAGAGCAGTTGGCTAAGAACAAGGAt........ | 29 | t | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TATGCAAGAGCAGTTGGCTAAGAACAAGG.......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGAGACCCCGTGAGGAAAGAAAGGCCGC.......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................CGTGAGACCCCGTGAGGAAAGAAAGGC............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................AAGGTCAGACTGGGCTACCTCGTGA................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGGCTAAGAACAAGG.......... | 15 | 12 | 0.08 | 0.08 | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGAAATCTTGGGCTGCCTACCTGTGAAGCCAGCACTAGAAAGGCTGAGGCACGACCCATTCAAGGTCAGACTGGGCTACCTCGTGAGACCCCGTGAGGAAAGAAAGGCCGCTTTAAGAAGTGAGGGGCAAGCAAGGTTCTTGGTCATTTCTGTGGTCAGTAATGACAAAGCTCCTGCACTGGGGCACTCCTTCCTCTAGGCCCGCCAAGCTATGCAAGAGCAGTTGGCTAAGAACAAGGAACTGACCCA ..................................................(((((((((.((.......)).)))).....))))).....((....))....................................................................................................................................................... ..................................................51.....................................................106.............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................AGGTTCTTGGTCATTTCTGTGGTCAGTA........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GCTAAGAACAAGGAacaa......... | 18 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGACAAAGCTCCTGgacc......................................................................... | 18 | gacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |