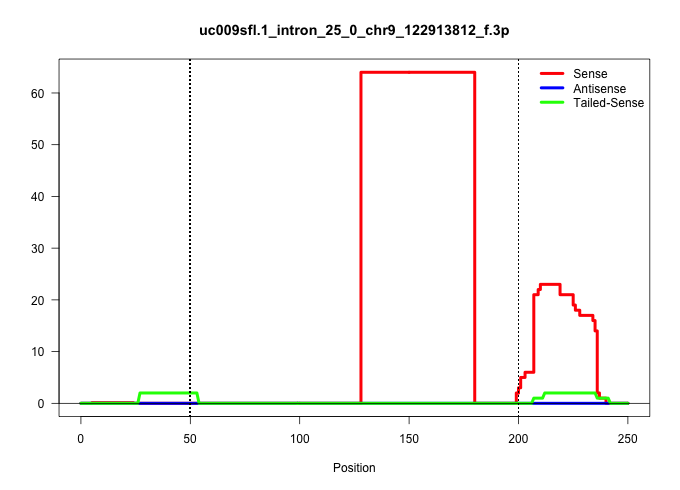
| Gene: Kif15 | ID: uc009sfl.1_intron_25_0_chr9_122913812_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| GGGAATTGAACTCAGGATGTCTAGAAGAGCAGTCAGCGCTCTTAACCACTGAGCCGTTTCTGGAGCCCAGGAAAAAACGTTTTAAATGTTTAAGTATTTCCTTTCCTAACTTTAAATTTGTATAGGACTATAGTCATGATTGTGAGCTGAATGGCATAGTCAGAAATAAACCTTTTCTTTGGGACATTGCTCCCTGACAGAGGGACATGCTCTGTGAAGATCTGGCCCATGCCACTGAGCAGCTGAACAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................TATAGTCATGATTGTGAGCTGAATGGCATAGTCAGAAATAAACCTTTTCTTT...................................................................... | 52 | 1 | 64.00 | 64.00 | 64.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCTCTGTGAAGATCTGGCCCATGCCACT.............. | 29 | 1 | 12.00 | 12.00 | - | 3.00 | 4.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GGGACATGCTCTGTGAAG............................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GAGGGACATGCTCTGTGAAGATCTGG......................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...........................AGCAGTCAGCGCTCTTAACCACTGAGa.................................................................................................................................................................................................... | 27 | a | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCTCTGTGAAGATCTGGCCCATGCCAC............... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................GACATGCTCTGTGAAGATCTGGCCC...................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AGGGACATGCTCTGTGAAGATCTGGC........................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCTCTGTGAAGATCTGGCCCATGCCA................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TCTGTGAAGATCTGGCCCATGCCACTGAGC.......... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................................................CTCTGTGAAGATCTGGCCCATGCCACTG............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCTCTGTGAAGATCTGGCCCATGCCAat.............. | 29 | at | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGTGAAGATCTGGCCCATGCCACTGAGCAt........ | 30 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TTGAACTCAGGATGTCTAG.................................................................................................................................................................................................................................. | 19 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |
| GGGAATTGAACTCAGGATGTCTAGAAGAGCAGTCAGCGCTCTTAACCACTGAGCCGTTTCTGGAGCCCAGGAAAAAACGTTTTAAATGTTTAAGTATTTCCTTTCCTAACTTTAAATTTGTATAGGACTATAGTCATGATTGTGAGCTGAATGGCATAGTCAGAAATAAACCTTTTCTTTGGGACATTGCTCCCTGACAGAGGGACATGCTCTGTGAAGATCTGGCCCATGCCACTGAGCAGCTGAACAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................AGGGACATGCTCTGggct.................................... | 18 | ggct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........GGATGTCTAGAAGAagct.............................................................................................................................................................................................................................. | 18 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................AGAGCAGTCAGCGCTCTTAACCACTGAGcg..................................................................................................................................................................................................... | 30 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TCCCTGACAGAGGGAaga............................................. | 18 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................AGCAGTCAGCGCTCTTAACCACTaaa........................................................................................................................................................................................................ | 26 | aaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |