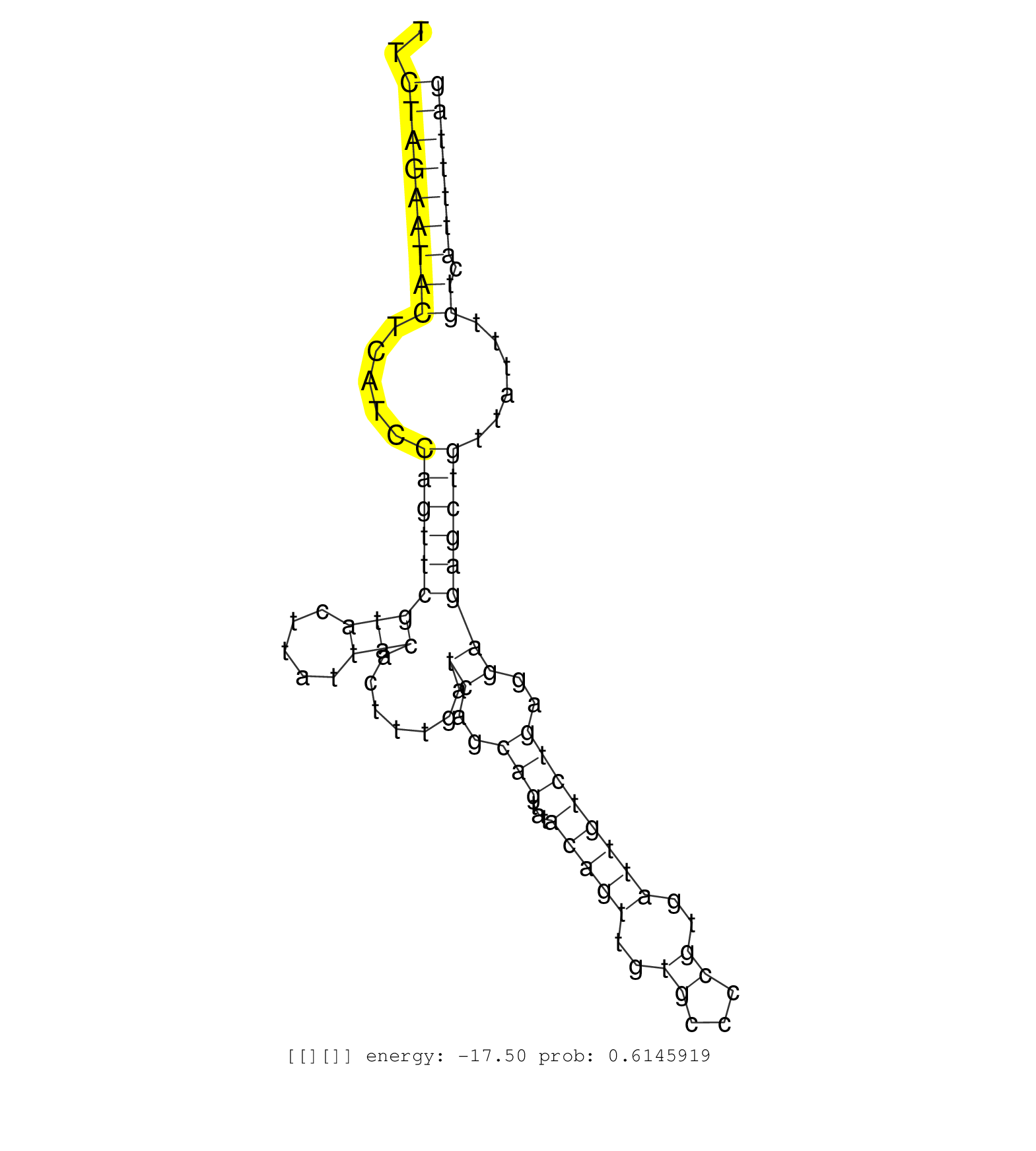

| Gene: AK006734 | ID: uc009seg.1_intron_1_0_chr9_121848620_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTATGAAGTTGCTAGATCATAAAGAGTGCATTTGGTTAGTCTTACTATCTGCTCTACAAACGTTTTCTAGAATACTCATCCAGTTCGTACTTATTACACTTTGATCAGCAGTATACAGTTGTGCCCCGTGATTGTCTGAGGAGAGCTGTTATTTGTCATTTTAGGAACCAGTGATATTTGAAGATGAGGCTGTGTACTTCACGGAGCAGGAATT ......................................................................................................(((((((((.....(((((((((.....))).......((..(((...(((((..((...))..))))))))..))))))))......)).))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................TAAAGAGTGCATTTGGTTAGTCTTACTATCTGCTCTACAAACGTTTTCTAGA............................................................................................................................................... | 52 | 1 | 90.00 | 90.00 | 90.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGTGTAC................. | 26 | 1 | 16.00 | 16.00 | - | 5.00 | 4.00 | 3.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................................................TGAAGATGAGGCTGTGTACTTCACGGA......... | 27 | 1 | 11.00 | 11.00 | - | 5.00 | - | - | - | 3.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGT................... | 26 | 1 | 7.00 | 7.00 | - | 5.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGTGTACT................ | 27 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGTGTA.................. | 25 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................GTGATATTTGAAGATGAGGCTGTGTAC................. | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TTTGAAGATGAGGCTGTGTACTTCACGGA......... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGggt................... | 26 | ggt | 3.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTAC................. | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TTGAAGATGAGGCTGTGTACTTCACGG.......... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAACCAGTGATATTTGAAGATGAGGC........................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTA.................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTAt................. | 28 | t | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAACCAGTGATATTTGAAGATGAGG......................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CTGTGTACTTCACGGAGC....... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TTTGAAGATGAGGCTGTGTACTTCACGG.......... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGT..................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTACTT............... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATGAGGCTGTGTACTTCACGGAGCAGGAATT | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGTGTAa................. | 26 | a | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGggta.................. | 25 | ggta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGATATTTGAAGATGAGGCTGTGgaa................. | 26 | gaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................TGAAGATGAGGCTGTGTACTT............... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTACT................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GATATTTGAAGATGAGGCTGTGTACT................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGggtt.................. | 27 | ggtt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTG.................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATGAGGCTGTGTACTTCACGGAGCAG..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................TTTGAAGATGAGGCTGTGTACTTCAC............ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TTTGAAGATGAGGCTGTGTACTTCACGGg......... | 29 | g | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATTTGAAGATGAGGCTGTGTACTTCAC............ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGTGATATTTGAAGATGAGGCTGTGTAa................. | 28 | a | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TTGAAGATGAGGCTGTGTACTTCACGGAGC....... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAACCAGTGATATTTGAAGATGAGGCTG...................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTATGAAGTTGCTAGATCATAAAGAGTGCATTTGGTTAGTCTTACTATCTGCTCTACAAACGTTTTCTAGAATACTCATCCAGTTCGTACTTATTACACTTTGATCAGCAGTATACAGTTGTGCCCCGTGATTGTCTGAGGAGAGCTGTTATTTGTCATTTTAGGAACCAGTGATATTTGAAGATGAGGCTGTGTACTTCACGGAGCAGGAATT ......................................................................................................(((((((((.....(((((((((.....))).......((..(((...(((((..((...))..))))))))..))))))))......)).))))))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) |
|---|