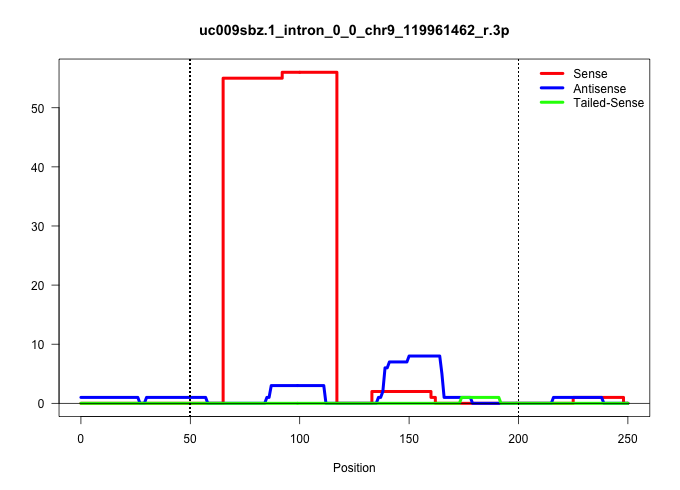
| Gene: Cx3cr1 | ID: uc009sbz.1_intron_0_0_chr9_119961462_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(2) PIWI.mut |
(12) TESTES |
| CCTTTCAGTGTTTTCTCCCGCTTGCTGCATGCAGCCAGTGAGAACCGCGATCCTCTAAGACTCACGTGATCTGGTTTGCTGCATACAGCCAGTGAGAACTGCGATCCTCTAAGACTCACGTGGACCTGCTTACTGCATGCAGCCAGTGAGAACTACAATCCTTTAAGGCTCACGTGATCTGGTTTCTCCTTCCCCTCCAGGACCTCACCATGTCCACCTCCTTCCCTGAACTGGATCTAGAGAATTTTGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................GTGATCTGGTTTGCTGCATACAGCCAGTGAGAACTGCGATCCTCTAAGACTC..................................................................................................................................... | 52 | 1 | 55.00 | 55.00 | 55.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGCATGCAGCCAGTGAGAACTACAATCCT........................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................TGAGAACTGCGATCCTCTAAGACTC..................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CTGAACTGGATCTAGAGAATTTT.. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGATCTGGTTTCTCagcg.......................................................... | 18 | agcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................TGCATGCAGCCAGTGAGAACTACAATC.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CCTTTCAGTGTTTTCTCCCGCTTGCTGCATGCAGCCAGTGAGAACCGCGATCCTCTAAGACTCACGTGATCTGGTTTGCTGCATACAGCCAGTGAGAACTGCGATCCTCTAAGACTCACGTGGACCTGCTTACTGCATGCAGCCAGTGAGAACTACAATCCTTTAAGGCTCACGTGATCTGGTTTCTCCTTCCCCTCCAGGACCTCACCATGTCCACCTCCTTCCCTGAACTGGATCTAGAGAATTTTGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................CAGCCAGTGAGAACTACAATCCTTTAA.................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................GCCAGTGAGAACTGCGATCCTCTAA.......................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ........................................................CTCACGTGATCTGGTTTGCTGCAccct....................................................................................................................................................................... | 27 | ccct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................AACTACAATCCTTTAAGGCTCACGTGATC....................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GCAGCCAGTGAGAACCGCGATCCTCTAA................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................CAGCCAGTGAGAACTGCGATCCTCTAA.......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................CTGCATACAGCCAGTGAaggt........................................................................................................................................................... | 21 | aggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................CAGCCAGTGAGAACTACAATCCTTTA..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CCTCCTTCCCTGAACTGGATCTA........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................GCCAGTGAGAACTACAATCCTTTAA.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ATGCAGCCAGTGAGAACTACAATCCTTTA..................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................GCAGCCAGTGAGAACTACAATCCTTTA..................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |