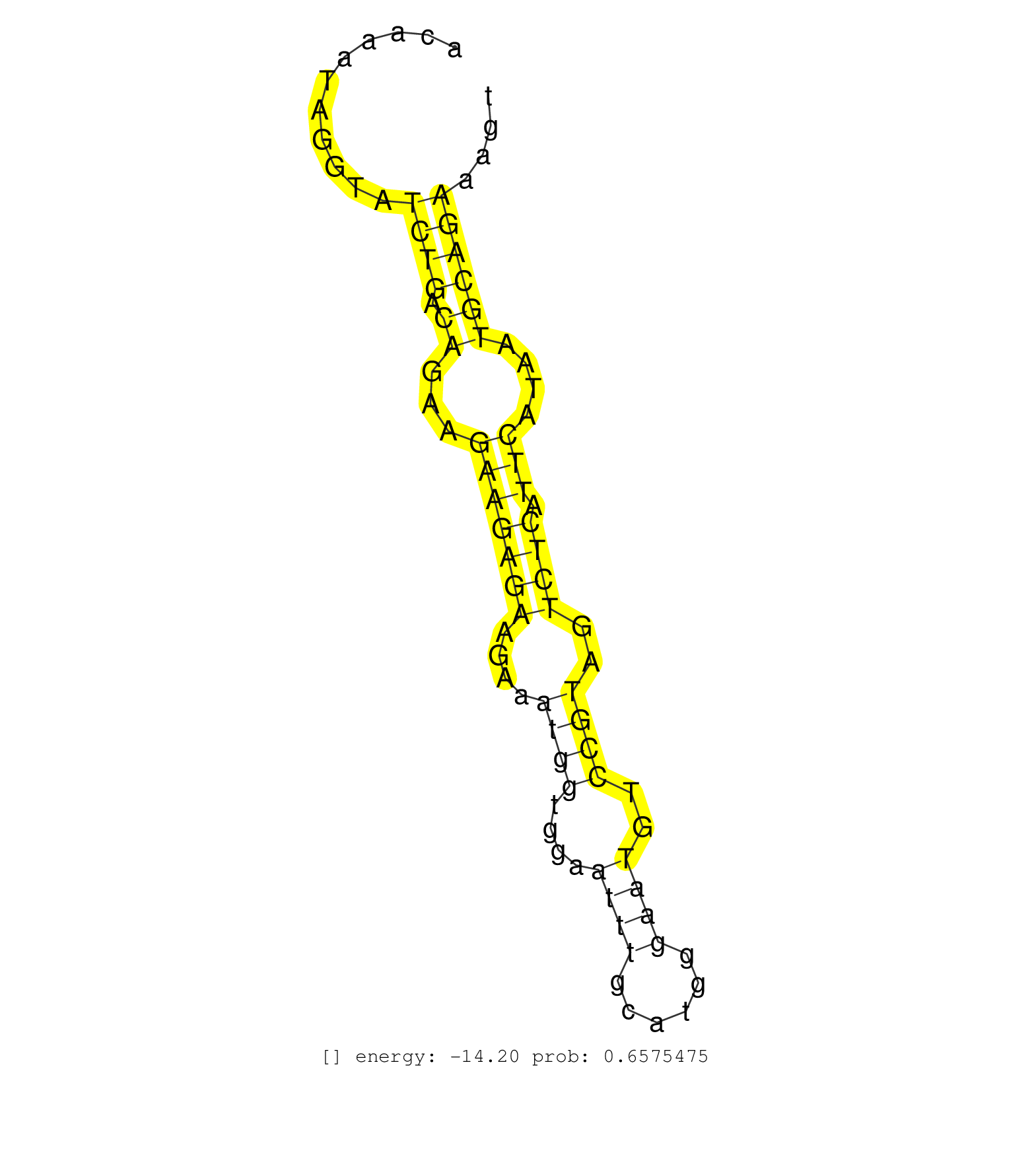
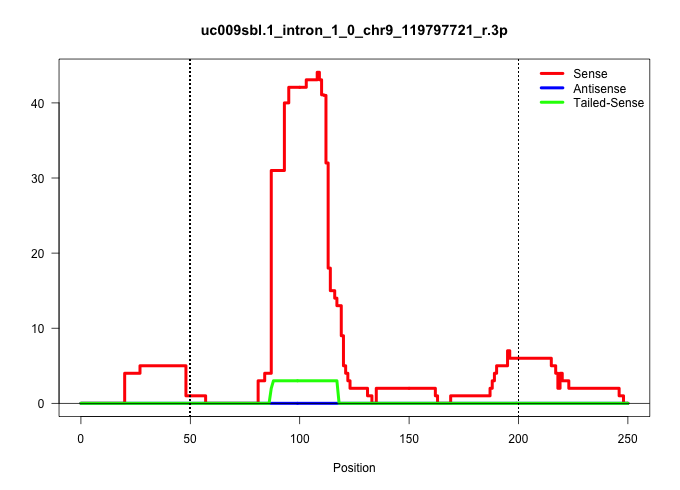
| Gene: AK076661 | ID: uc009sbl.1_intron_1_0_chr9_119797721_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| TTAGTAATACAAAGAATGGTTACAAGCTAACAGAATACACTAAAACACAATAGAAAAAAAAAAAAAAAAACGAGCAAAGGGTACAAATAGGTATCTGACAGAAGAAGAGAAGAAATGGTGGAATTTGCATGGGAATGTCCGTAGTCTCATTCATAATGCAGAAAGTACATGTTGAAGGGAACACCACTTTTCTCCTACAGCTCGGAAAAGACCGGACCGTTTAATGAGATGCCGTGCTCAGGCGATGTTT .............................................................................................((((.((...(((((((....((((....((((......))))..))))..)))).)))....))))))........................................................................................ ..................................................................................83.................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAGA......................................................................................................................................... | 26 | 1 | 14.00 | 14.00 | 4.00 | 2.00 | 1.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAG.......................................................................................................................................... | 25 | 1 | 7.00 | 7.00 | 2.00 | - | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TCTGACAGAAGAAGAGAAGAAATGGT................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TACAAGCTAACAGAATACACTAAAACAC.......................................................................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TCTGACAGAAGAAGAGAAGAAATGGTG.................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAGAA........................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TACAAATAGGTATCTGACAGAAGAAGAGAAG.......................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAGAAATGt.................................................................................................................................... | 31 | t | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................AGGTATCTGACAGAAGAAGAGAAGAAATGt.................................................................................................................................... | 30 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCTCCTACAGCTCGGAAAAGACCGGACC................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACAGAAGAAGAGAAGAAATGGTGGA................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TTCTCCTACAGCTCGGAAAAGACCGGACC................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................GAAGAGAAGAAATGGTGGAATTTGCATG....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAGAAATG..................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................TGTCCGTAGTCTCATTCATAATGCAGA........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TTTAATGAGATGCCGTGCTCAGGCGATGT.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................AAATAGGTATCTGACAGAAGAAGAGA............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................GAAGAAATGGTGGAATTTGCATGGG..................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAGAAGAAAT...................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTTTCTCCTACAGCTCGGAAAAGACCGG................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................TACAAATAGGTATCTGACAGAAGAAGAGA............................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................TCTGACAGAAGAAGAGAAGAAATGGTGG................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TTTAATGAGATGCCGTGCTCAGGCGAT.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................TAGGTATCTGACAGAAGAAGAG............................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TTTCTCCTACAGCTCGGAAAAGACCGGAC................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TACAGCTCGGAAAAGACCGGACCGTTTA........................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGACAGAAGAAGAGAAGAAATGGTGGAA............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TAACAGAATACACTAAAACACAATAGAAAA................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................TGTCCGTAGTCTCATTCATAATGCAGAA....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TACAGCTCGGAAAAGACCGGACCGT.............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGTTGAAGGGAACACCACTTTTCTCCT...................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................TGACAGAAGAAGAGAA........................................................................................................................................... | 16 | 13 | 0.08 | 0.08 | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - |
| TTAGTAATACAAAGAATGGTTACAAGCTAACAGAATACACTAAAACACAATAGAAAAAAAAAAAAAAAAACGAGCAAAGGGTACAAATAGGTATCTGACAGAAGAAGAGAAGAAATGGTGGAATTTGCATGGGAATGTCCGTAGTCTCATTCATAATGCAGAAAGTACATGTTGAAGGGAACACCACTTTTCTCCTACAGCTCGGAAAAGACCGGACCGTTTAATGAGATGCCGTGCTCAGGCGATGTTT .............................................................................................((((.((...(((((((....((((....((((......))))..))))..)))).)))....))))))........................................................................................ ..................................................................................83.................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|