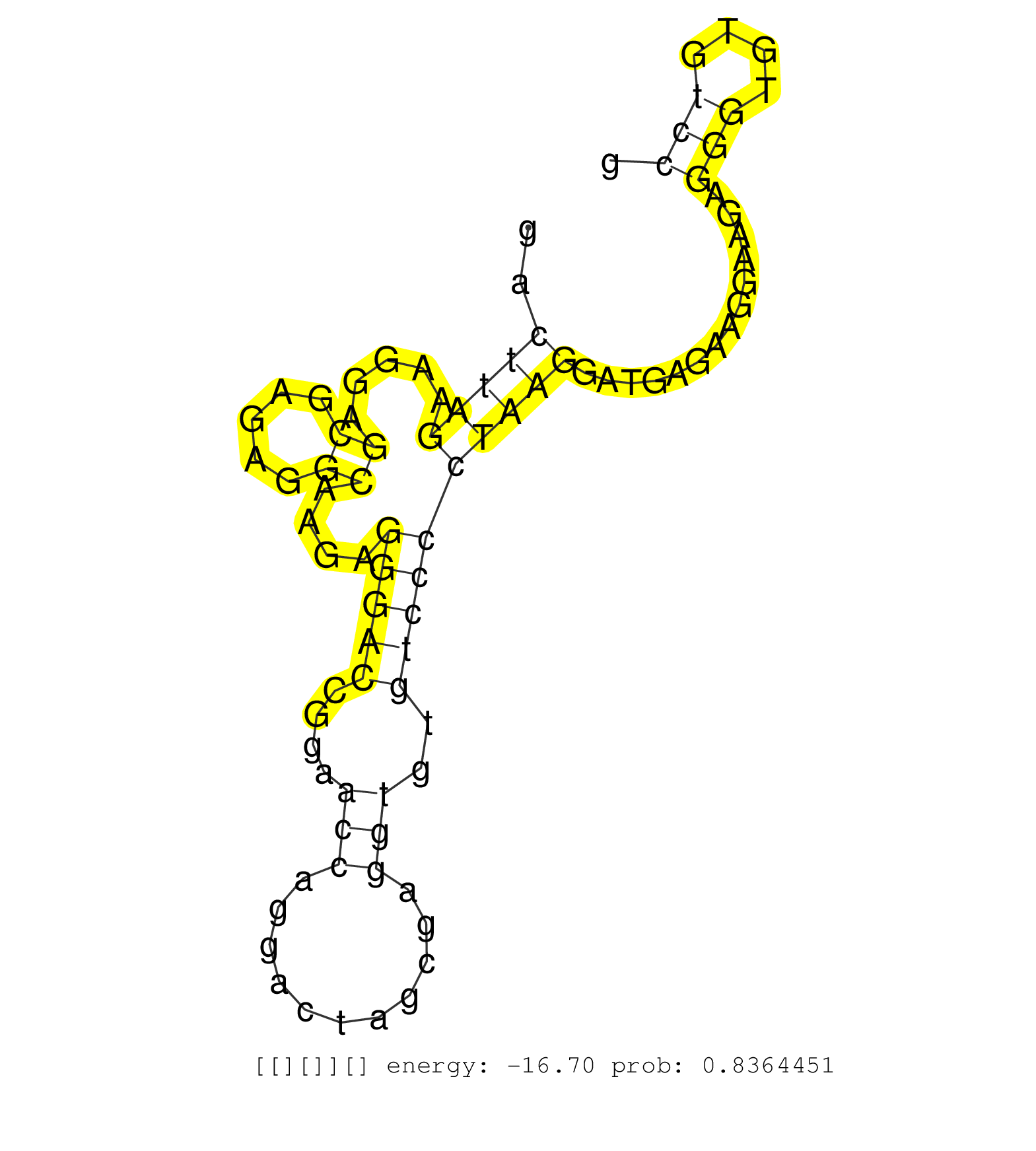

| Gene: Vill | ID: uc009sab.1_intron_9_0_chr9_118972502_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| CTTCTGGTCTTGGTCGAAGGAGCTAGATAGGAAAAAACACCCAGAGAAGAGTGAGCAAGAGGAGGGGCACGGGCGCGGGGGTGGAGCCTGGTGAATGGGCGGGACTTAGAAGGAGCGAGAGGCAAGAGGGACCGGAACCAGGACTAGCGAGGTGTGTCCCCTAAGGATGAGAAGGAAGAGGGTGTGTCCGGGACCCTGTGAGATGGGTGAGGAGTGATTCGGCCGGGTGGTCCTGAAGAACATCACTGACTCTCCGCTCCACATGCAGGTAAACTGGTGCAGGGAAACCTGGAGGTAGGCAAGCTTCACACCCAGCCT ........................................................................................................(((((.....((.....))....(((((....(((...........)))..))))))))))..............(((....)))................................................................................................................................. ......................................................................................................103....................................................................................190.............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................CGGGACCCTGTGAGATGGGTGAGGAGTG...................................................................................................... | 28 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGGATGAGAAGGAAGAGGGTGTG.................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................................TGAAGAACATCACTGACTCTCCGCT............................................................ | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGAAGGAAGAGGGTGTGTCCGGGA............................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TGAAGAACATCACTGACTCTCCGCTC........................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGTGAGATGGGTGAGGAGTGATTCGGt............................................................................................... | 27 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CGGGGGTGGAGCCTGcggc................................................................................................................................................................................................................................ | 19 | cggc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAAGGATGAGAAGGAAGAGGGTaa..................................................................................................................................... | 24 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGTGAGATGGGTGAGGAGTGATTCGG................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................AGGATGAGAAGGAAGAGGGTGTGTCC................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................TCACTGACTCTCCGCTCCACATGCAGGTg............................................... | 29 | g | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CTTGGTCGAAGGAGCTAGATAGGAAA............................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAGAAGGAAGAGGGTGTGTCCGGG.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................AGGCAAGAGGGACCGGAACCAG................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GGTGAGGAGTGATTCGG................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CTAAGGATGAGAAGGAAGAGGGTaa..................................................................................................................................... | 25 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................GTCCGGGACCCTGcatg.................................................................................................................... | 17 | catg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GAGGCAAGAGGGACCGGAACCAGGAC.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................................TCACTGACTCTCCGCTCCACATGCAGGgg............................................... | 29 | gg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGTGAGATGGGTGAGGAGTGATTCGt................................................................................................ | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................AACCTGGAGGTAGGCAAGCTTCACACCC..... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TGTGAGATGGGTGAGGAGTGATTCGGC............................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AGAAGGAGCGAGAGGCAAGAGGGACCG........................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TCCTGAAGAACATCACTGACTCTCCGC............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................GTAGGCAAGCTTCgg......... | 15 | gg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CTAAGGATGAGAAGGAAGAGGGTGT..................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................AGGAGCTAGATAGGAAAAAACACCCAGA................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AAGGATGAGAAGGAAGAGGGTGTGTCC................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................................................TGACTCTCCGCTCCACATGCAGGTga.............................................. | 26 | ga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CTAAGGATGAGAAGGAAGAG.......................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................AGGGACCGGAACCAGGACTAGCGAGGTGT................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................TGGAGGTAGGCAAGCTTCACACCCAGC.. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................TCACTGACTCTCCGCTCCACATGCAG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................................................GTGTCCGGGACCCTGT....................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .....................................................................CGGGCGCGGGGGTGG.......................................................................................................................................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTCTGGTCTTGGTCGAAGGAGCTAGATAGGAAAAAACACCCAGAGAAGAGTGAGCAAGAGGAGGGGCACGGGCGCGGGGGTGGAGCCTGGTGAATGGGCGGGACTTAGAAGGAGCGAGAGGCAAGAGGGACCGGAACCAGGACTAGCGAGGTGTGTCCCCTAAGGATGAGAAGGAAGAGGGTGTGTCCGGGACCCTGTGAGATGGGTGAGGAGTGATTCGGCCGGGTGGTCCTGAAGAACATCACTGACTCTCCGCTCCACATGCAGGTAAACTGGTGCAGGGAAACCTGGAGGTAGGCAAGCTTCACACCCAGCCT ........................................................................................................(((((.....((.....))....(((((....(((...........)))..))))))))))..............(((....)))................................................................................................................................. ......................................................................................................103....................................................................................190.............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................TCACTGACTCTCCGCTCCACATGCA................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |