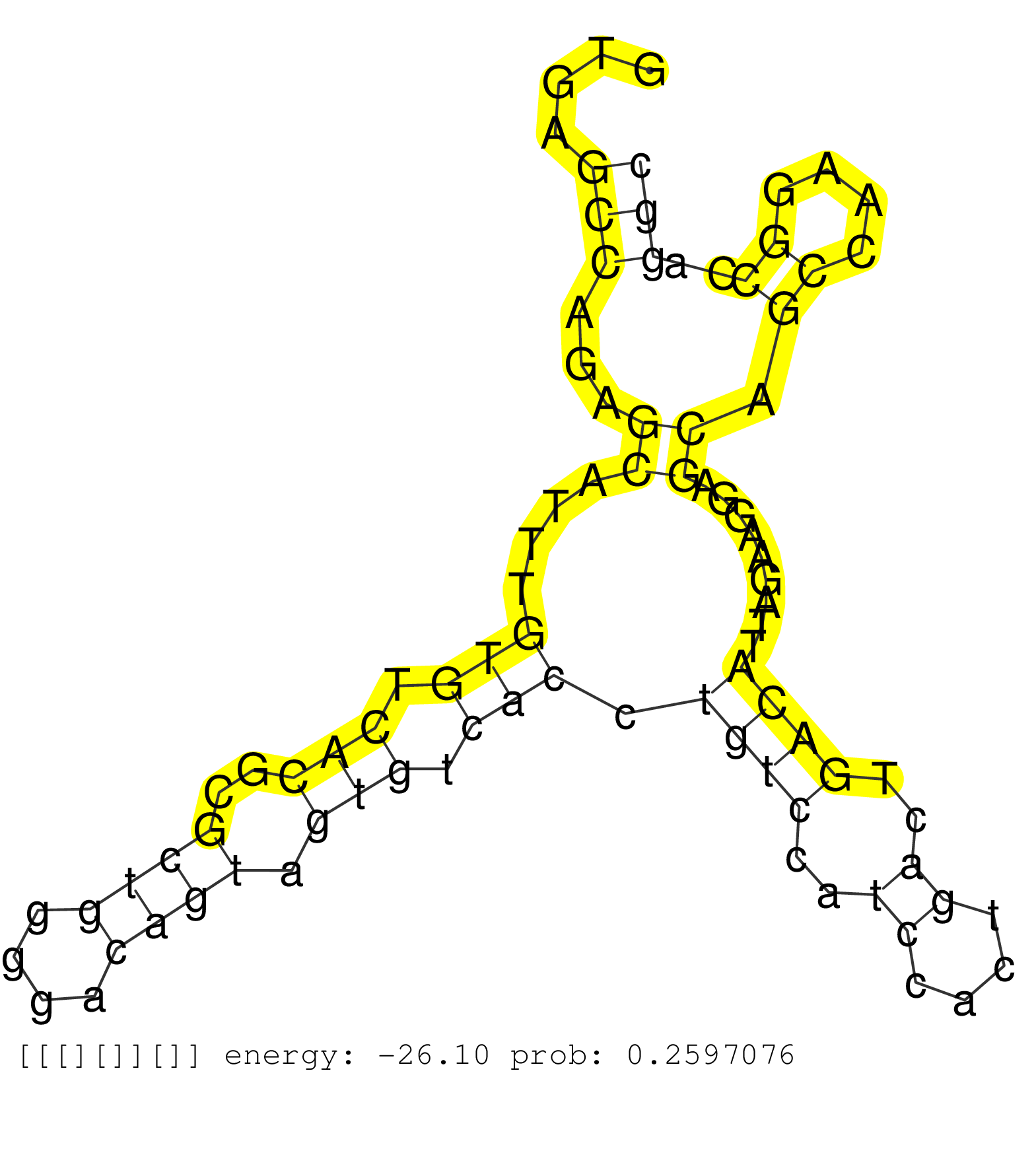

| Gene: 6620401K05Rik | ID: uc009rrt.1_intron_10_0_chr9_108971912_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(21) TESTES |
| AAGAAAAGACTCAAGCACTCAGTGAAAAGGAAAAGGAATTCTCCAGAAAGGTGAGCCAGAGCATTTGTGTCACGCGCTGGGGACAGTAGTGTCACCTGTCCATCCACTGACTGACATTAGAAGGAGCAGCCAAGGCCAGGCATGCTGACATATGCCTTTAACCTCAAGAGGCAGAGGCAGGTGGATCTCTGTGAGCTCTAGGCCAGCGTAGACTACACAGTGAATTCCAGGCCAGTCAAGTGAGAAACTT ......................................................(((...((....(((.(((..((((....)))).))).))).((((..((....))..)))).........)).((....))..)))............................................................................................................. ..................................................51........................................................................................141........................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................TGACATTAGAAGGAGCAGCCAAGGCC................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGAGCTCTAGGCCAGCGTAGACTAC.................................. | 27 | 1 | 7.00 | 7.00 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGAGCTCTAGGCCAGCGTAGACTACAC................................ | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAAAAGGAAAAGGAATTCTCCAGAA.......................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCACTGACTGACATTAGAAGGAGCAGC........................................................................................................................ | 28 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGAGCTCTAGGCCAGCGTAGACTA................................... | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACTGACATTAGAAGGAGCAGCCAAG.................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................TGTCCATCCACTGACTGACATTAGAAGG.............................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCACTGACTGACATTAGAAGGAGCAG......................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAAAGGAAAAGGAATTCTCCAGAAAGac...................................................................................................................................................................................................... | 28 | ac | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAAAAGGAAAAGGAATTCTCCAGAAAGc....................................................................................................................................................................................................... | 29 | c | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................AAAAGGAAAAGGAATTCTCCAGAAAGG....................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................TGACATTAGAAGGAGCAGCCAAGGCCAG............................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TGGATCTCTGTGAGCTCTAGGCCAGCGTA........................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .AGAAAAGACTCAAGCACTCAGTGAA................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................GTGAGCTCTAGGCCAGCGTAGACTAC.................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCCAGAGCATTTGTGTCACGCG.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................TGACATTAGAAGGAGCAGCCAAGGCCAGG.............................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGACATTAGAAGGAGCAGCCAAG.................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................ATCTCTGTGAGCTCTAGGCCAGCGTAGA...................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGAGCTCTAGGCCAGCGTAGA...................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGAGCTCTAGGCCAGCGTAGACTAt.................................. | 27 | t | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .AGAAAAGACTCAAGCACTCAGTGAAAAGGA........................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................................................TAGGCCAGCGTAGACTACACAGTGAA.......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAAAAGGAAAAGGAATTCTCCAGAAAGct...................................................................................................................................................................................................... | 30 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TACACAGTGAATTCCAGGCCAGTCAAGT......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................GAAAAGGAAAAGGAATTCTCCAGAAAGc....................................................................................................................................................................................................... | 28 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................TCCACTGACTGACATTAGAAGGAGCAGCC....................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAAAGGAAAAGGAATTCTCCAGAAAGaat..................................................................................................................................................................................................... | 29 | aat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGACATTAGAAGGAGCAGCCAAGGC.................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAAAGGAAAAGGAATTCTCCAGAAAGGgtc.................................................................................................................................................................................................... | 30 | gtc | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AGGAAAAGGAATTCTCCAGAAAGctc..................................................................................................................................................................................................... | 26 | ctc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGTGAGCTCTAGGCCAGCGTAGACTACA................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TACACAGTGAATTCCAGGCCAGTCAAGTG........ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGACTGACATTAGAAGGAGCAGCCAAGGC.................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AAAGACTCAAGCACTCAGTGAAAAGGA........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................TCTGTGAGCTCTAGGCCAGCGTA........................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TAGGCCAGCGTAGACTACACAGTGAATTCCAG.................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................TTAACCTCAAGAGGCAGAGGCAGGTG................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................ATCTCTGTGAGCTCTAGGCCAGC........................................... | 23 | 8 | 0.12 | 0.12 | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGAAAAGACTCAAGCACTCAGTGAAAAGGAAAAGGAATTCTCCAGAAAGGTGAGCCAGAGCATTTGTGTCACGCGCTGGGGACAGTAGTGTCACCTGTCCATCCACTGACTGACATTAGAAGGAGCAGCCAAGGCCAGGCATGCTGACATATGCCTTTAACCTCAAGAGGCAGAGGCAGGTGGATCTCTGTGAGCTCTAGGCCAGCGTAGACTACACAGTGAATTCCAGGCCAGTCAAGTGAGAAACTT ......................................................(((...((....(((.(((..((((....)))).))).))).((((..((....))..)))).........)).((....))..)))............................................................................................................. ..................................................51........................................................................................141........................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................GAGGCAGAGGCAGGTGGATCTCTGggaa........................................................... | 28 | ggaa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |