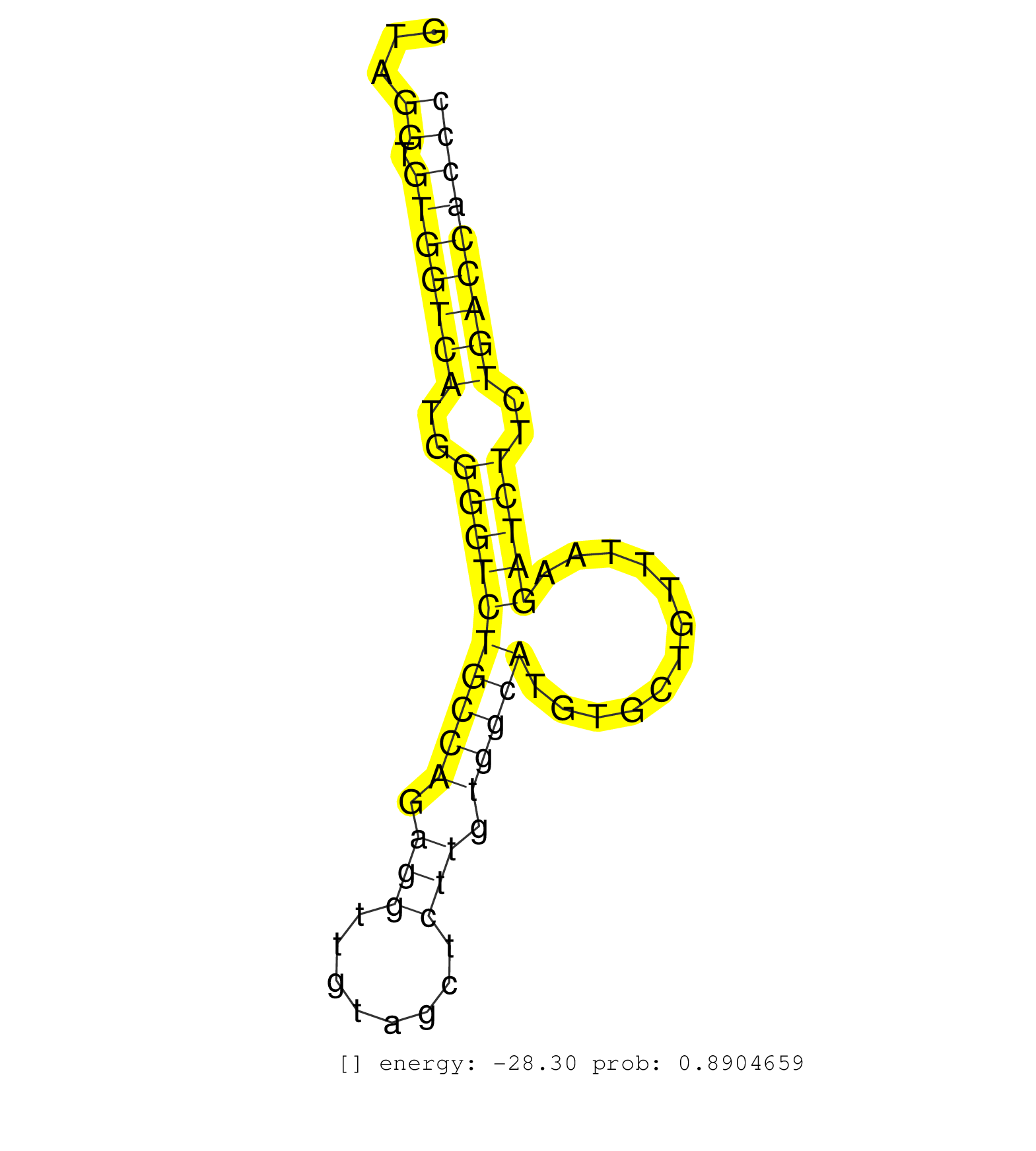
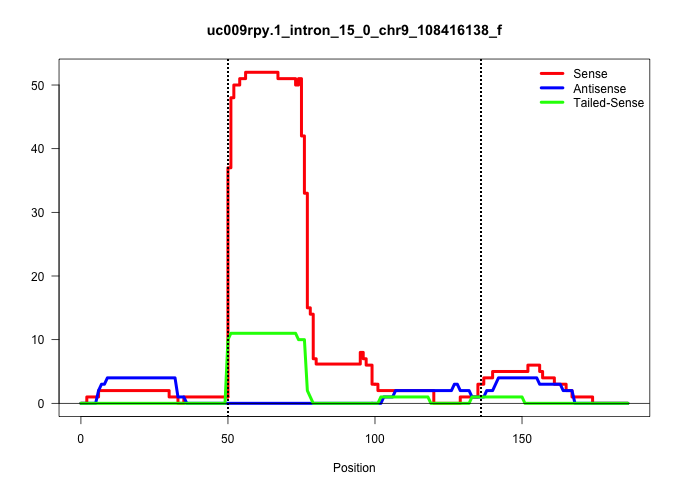
| Gene: Qars | ID: uc009rpy.1_intron_15_0_chr9_108416138_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(3) PIWI.mut |
(29) TESTES |
| GTTGTCTCAAAGCGCAAGATTCTCCAGCTTGTAGCAGCTGGTGCTGTTCGGTAGGTGTGGTCATGGGGTCTGCCAGAGGTTGTAGCTCTTGTGGCATGTGCTGTTTAAGATCTTCTGACCACCCTCTTATCCAAAGGGACTGGGACGATCCACGGCTCTTCACACTCACTGCCCTACGACGACGGG .....................................................((.(((((((..((((((((((.(((........))).)))))............)))))..))))))))).............................................................. ..................................................51.......................................................................124............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAG.............................................................................................................. | 26 | 1 | 10.00 | 10.00 | 2.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCA............................................................................................................... | 25 | 1 | 10.00 | 10.00 | 1.00 | 1.00 | 1.00 | - | - | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAGA............................................................................................................. | 27 | 1 | 9.00 | 9.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTGTGGTCATGGGGTCTGCCAGA............................................................................................................. | 26 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAGt............................................................................................................. | 27 | t | 5.00 | 10.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGGTTGTAGCTCTTGTGGCATGT....................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAGAGG........................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTGTGGTCATGGGGTCTGCCAGAGG........................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCC................................................................................................................ | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGGTTGTAGCTCTTGTGGCATGTGC..................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................ATGTGCTGTTTAAGATCTTCTGACC.................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................CGGCTCTTCACACTCACTGCCC............ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGGGACGATCCACGGCTCTTCACACTC................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTGTGGTCATGGGGTCTGCCAGAG............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGTGTGGTCATGGGGTCTGCCAGA............................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCAGCTTGTAGCAGCTGGTGCTGTTCGG....................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................AGGTTGTAGCTCTTGTGGCATGT....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGC................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AAGGGACTGGGACGtgtg................................... | 18 | tgtg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GTTTAAGATCTTCgtta................................................................... | 17 | gtta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGTGGTCATGGGGTCTGCCAGAGG........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................AGGTTGTAGCTCTTGTGGCA.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAGAGc........................................................................................................... | 29 | c | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGGTTGTAGCTCTTGTGGCAT......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TCAAAGCGCAAGATTCTCCAGCTTGTA......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GACTGGGACGATCCACGGCTCTTCACAC..................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGTGTGGTCATGGGGTCTGCCAGAGGT.......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGACTGGGACGATCCACGGCT............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................TCCAGCTTGTAGCAGCTGGTGCTGTTCG........................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................AGCTGGTGCTGTTCGGT...................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGTGTGGTCATGGGGTCTGCCAGAGG........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGTCTCAAAGCGCAAGATTCTCCAGCTT............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCcga............................................................................................................. | 27 | cga | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGG....................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTcca................................................................................................................ | 24 | cca | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCAAAGGGACTGGGACGATCCACGGC.............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAtt............................................................................................................. | 27 | tt | 1.00 | 10.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGACTGGGACGATCCACGGCTCTTC......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................GTGGTCATGGGGTCTGCCAG.............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGTGTGGTCATGGGGTCTGCCAGAa............................................................................................................ | 27 | a | 1.00 | 8.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTGTGGTCATGGGGTCTGCCAaa............................................................................................................. | 27 | aa | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGTAGCTCTTGTGGC........................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTTGTCTCAAAGCGCAAGATTCTCCAGCTTGTAGCAGCTGGTGCTGTTCGGTAGGTGTGGTCATGGGGTCTGCCAGAGGTTGTAGCTCTTGTGGCATGTGCTGTTTAAGATCTTCTGACCACCCTCTTATCCAAAGGGACTGGGACGATCCACGGCTCTTCACACTCACTGCCCTACGACGACGGG .....................................................((.(((((((..((((((((((.(((........))).)))))............)))))..))))))))).............................................................. ..................................................51.......................................................................124............................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................AGATCTTCTGACCACCCTCTTATCCA..................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GGACGATCCACGGCTCTTCAtagt........................ | 24 | tagt | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TCAAAGCGCAAGATTCTCCAGCTTGTA......................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACTGGGACGATCCACGGCTCTTCACA...................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTTAAGATCTTCTGACCACCCTCTTA......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TATCCAAAGGGACTGGGACGATCCACGGC.............................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTCTGACCACCCTCTTATCCAAAGGGA............................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CAAAGCGCAAGATTCTCCAGCTTGTAtt......................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CAAAGCGCAAGATTCTCCAGCTTGTA......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GACGATCCACGGagct............................... | 16 | agct | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CTGGGACGATCCACGGCTCTTCAa........................ | 24 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGGACGATCCACGGCTCTTCAcgtc........................ | 25 | cgtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........AAGCGCAAGATTCTCCAGCTTGTAGCA...................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCAAAGCGCAAGATTCTCCAGCTTGTAtt......................................................................................................................................................... | 29 | tt | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GGGACGATCCACGGCTCTTCACACTCA.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GGACGATCCACGGCTCTTCACACTCA.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |