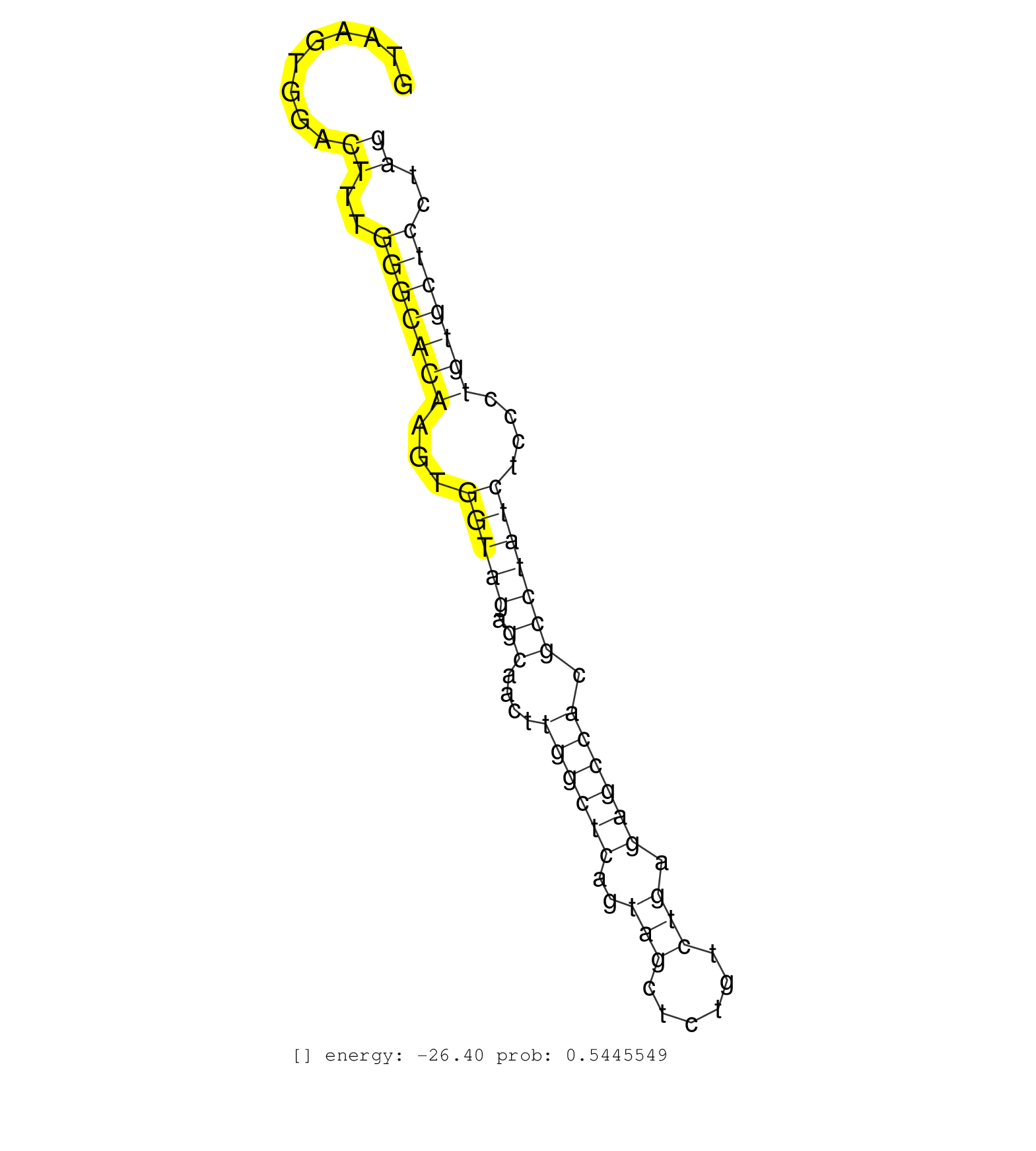

| Gene: Ifrd2 | ID: uc009rmd.1_intron_5_0_chr9_107493178_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
| GTGCTTCTGCTCTTGGCTTGGGTTGCTATGTGGCTGCCACCGACGTCCAGGTAAGTGGACTTTGGGCACAAGTGGTAGAGCAACTTGGCTCAGTAGCTCTGTCTGAGAGCCACGCCTATCTCCCTGTGCTCCTAGGACCTGGTCTCTTGCTTGGCCTGCTTGGAAGGTGTTTTCAGCTGGTCCTG ...........................................................((..(((((((...(((((.((....((((((..(((......))).)))))).)))))))....)))))))..)).................................................. ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGGACTTTGGGCACAAGTGGT............................................................................................................. | 26 | 1 | 26.00 | 26.00 | 6.00 | 7.00 | 2.00 | 2.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 |
| ..................................................GTAAGTGGACTTTGGGCACAAGTGGTA............................................................................................................ | 27 | 1 | 22.00 | 22.00 | 8.00 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - |
| ..................................................GTAAGTGGACTTTGGGCACAAGTGG.............................................................................................................. | 25 | 1 | 14.00 | 14.00 | 3.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - |
| ..................................................GTAAGTGGACTTTGGGCACAAGTGGTAt........................................................................................................... | 28 | t | 2.00 | 22.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGGACTTTGGGCACAAGTGGTAGAGC........................................................................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTGGACTTTGGGCACAAGTGGTA............................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................AAGTGGACTTTGGGCACAAGTGGTAGA.......................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGTGGCTGCCACCGACGTCCAGGacct.................................................................................................................................. | 27 | acct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGGACTTTGGGCACAAGTGGTAGA.......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGGACTTTGGGCACAAGTGGTAG........................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CTTGGCCTGCTTGGAAGGTGTTTTCA.......... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTTGGCCTGCTTGGAAGGTGTTTT............ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGGTAGAGCAACggac................................................................................................. | 16 | ggac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................TGCTATGTGGCTGCCACCGACGTCC......................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGGCACAAGTGGTAGAGCAACTTGGCT............................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGCTATGTGGCTGCCACCGACGTCCA........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGTGGCTGCCACCGACGTCCAGGacaa.................................................................................................................................. | 27 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCTCTTGGCTTGGGTTGCTATGTGG........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGGACTTTGGGCACAAGTGGT............................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCTCTTGGCTTGGGTTGCTATGTGGC....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGACTTTGGGCACAAGTGGTAGA.......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TCTGCTCTTGGCTTGGGTTGCTATGT.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................TGCTCCTAGGACCTGGTCTCTTGCTTG................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GGACTTTGGGCACAAGTGGT............................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........TCTTGGCTTGGGTTGCTATGTGGCTGC.................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTTGGCCTGCTTGGAAGGTGTTTTC........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................GCTATGTGGCTGCCACCGACGTCCcg....................................................................................................................................... | 26 | cg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TTTGGGCACAAGTGGTAGAGCAACTTG.................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGACTTTGGGCACAAG................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTGGACTTTGGGCACAAGTGtt............................................................................................................. | 26 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTTGGCCTGCTTGGAAGGTGTT.............. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GTGCTTCTGCTCTTGGCTTGGGTTGCTATGTGGCTGCCACCGACGTCCAGGTAAGTGGACTTTGGGCACAAGTGGTAGAGCAACTTGGCTCAGTAGCTCTGTCTGAGAGCCACGCCTATCTCCCTGTGCTCCTAGGACCTGGTCTCTTGCTTGGCCTGCTTGGAAGGTGTTTTCAGCTGGTCCTG ...........................................................((..(((((((...(((((.((....((((((..(((......))).)))))).)))))))....)))))))..)).................................................. ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|