
| Gene: Dock3 | ID: uc009rkv.1_intron_10_0_chr9_106815314_r | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(2) PIWI.ip |
(20) TESTES |
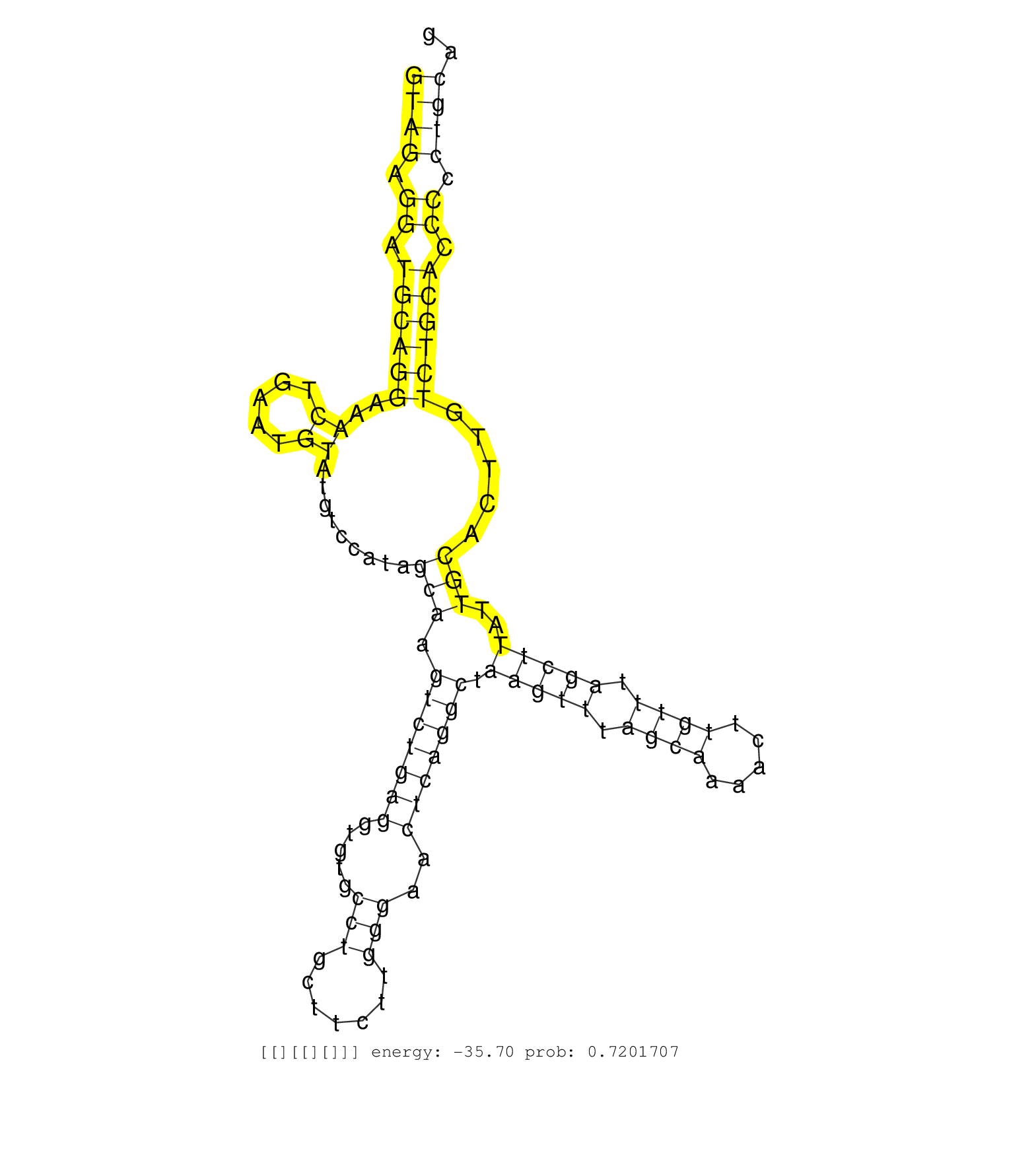
| ATGATAGACCTTTCCACAAAGGCCCCAAGGACAAAGACAATGAATTCAAGGTAGAGGATGCAGGAAACTGAATGTATGTCCATAGCAAGTCTGAGGTGTGCCTGCTTCTTGGGAACTCAGGCTAAGTTTAGCAAAACTTGTTTAGCTTATTGCACTTGTCTGCACCCCCTGCAGAGCCTGTGGATTGAGCGCACCACGCTGACACTGACCCACAGTTTGCCTGG ..................................................((((.((.((((((..((.....)).........(((.(((((((.....(((.......)))..))))))).(((((.((((.....)))).)))))..))).....)))))).)).)))).................................................... ..................................................51.........................................................................................................................174................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGAGGATGCAGGAAACTGAATGTA.................................................................................................................................................... | 26 | 1 | 14.00 | 14.00 | 3.00 | 4.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................TATTGCACTTGTCTGCctgt......................................................... | 20 | ctgt | 5.00 | 0.00 | - | - | - | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCccgt............................................................ | 17 | ccgt | 5.00 | 0.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CCAAGGACAAAGACAATGAATTCAAGagc........................................................................................................................................................................... | 29 | agc | 5.00 | 1.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCTcggc........................................................... | 18 | cggc | 5.00 | 0.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGAGGATGCAGGAAACTGAATGT..................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCTcg............................................................. | 16 | cg | 4.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCcagg............................................................ | 17 | cagg | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGAGGATGCAGGAAACTGAATGTATGT................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGAGGATGCAGGAAACTGAATGTATG.................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCTcgg............................................................ | 17 | cgg | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGGATTGAGCGCACCACGCTGACACTGA................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTAGAGGATGCAGGAAACTGAATGTATGT................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CCCCAAGGACAAAGACAATGAATTC................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AGGACAAAGACAATGAATTCAAGagcc.......................................................................................................................................................................... | 27 | agcc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CCCCAAGGACAAAGACAATGAATTCAAG.............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................GCCCCAAGGACAAAGACAATGAATTCAAGag............................................................................................................................................................................ | 31 | ag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCgg.............................................................. | 15 | gg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAAGGACAAAGACAATGAATTCAAGagc........................................................................................................................................................................... | 28 | agc | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CACAAAGGCCCCAAGGACAAAGA........................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCCAAGGACAAAGACAATGAATTCAAGagc........................................................................................................................................................................... | 30 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........CTTTCCACAAAGGCCCtaac................................................................................................................................................................................................... | 20 | taac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAGAGGATGCAGGAAACTGAATGTAT................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCCAAGGACAAAGACAATGAATTCAAGa............................................................................................................................................................................. | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGAGGATGCAGGAAACTGAATGTATGTC................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CAAGGACAAAGACAATGAATTCAAG.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCTGgcct.......................................................... | 19 | gcct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CCAAGGACAAAGACAATGAATTCAAGag............................................................................................................................................................................ | 28 | ag | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TGTATGTCCATAGCAAGTCTGAGGTGT............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................TATTGCACTTGTCctgg............................................................ | 17 | ctgg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCcgg............................................................. | 16 | cgg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTGCACTTGTCctg............................................................. | 16 | ctg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CCAAGGACAAAGACAATGAATTCAAG.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................AGACAATGAATTCAAG.............................................................................................................................................................................. | 16 | 5 | 0.20 | 0.20 | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATGATAGACCTTTCCACAAAGGCCCCAAGGACAAAGACAATGAATTCAAGGTAGAGGATGCAGGAAACTGAATGTATGTCCATAGCAAGTCTGAGGTGTGCCTGCTTCTTGGGAACTCAGGCTAAGTTTAGCAAAACTTGTTTAGCTTATTGCACTTGTCTGCACCCCCTGCAGAGCCTGTGGATTGAGCGCACCACGCTGACACTGACCCACAGTTTGCCTGG ..................................................((((.((.((((((..((.....)).........(((.(((((((.....(((.......)))..))))))).(((((.((((.....)))).)))))..))).....)))))).)).)))).................................................... ..................................................51.........................................................................................................................174................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................CTGAATGTATGTCCATAGCAAGTCTGA.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GCAGGAAACTGAATGTATGTCCATAGCAA........................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |