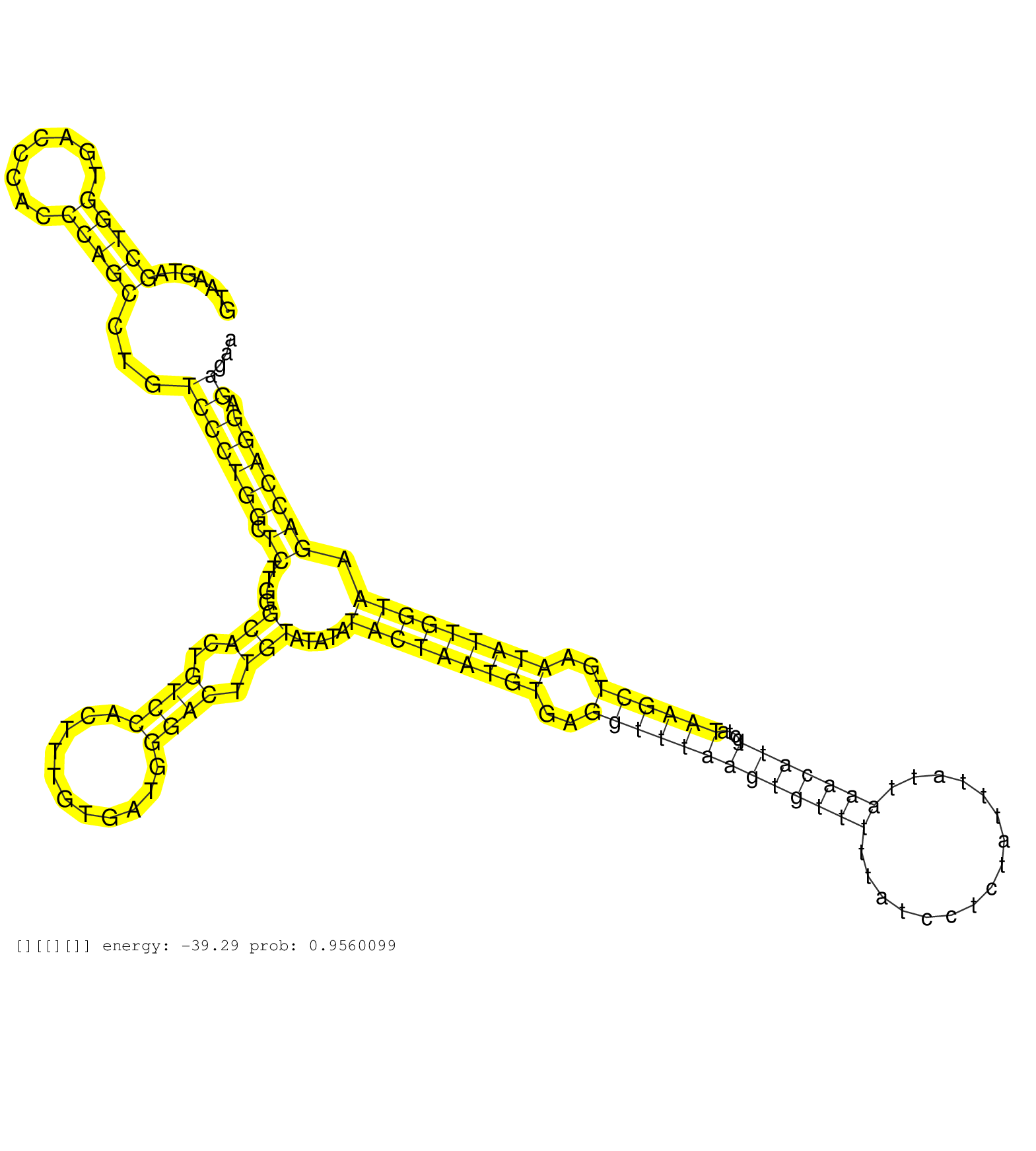
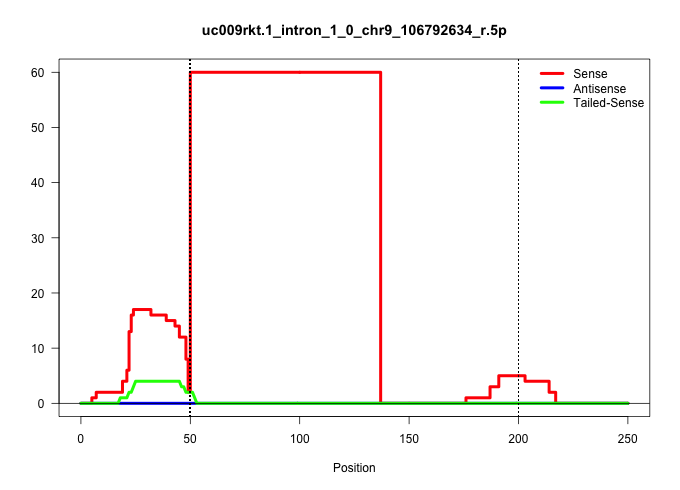
| Gene: Armet | ID: uc009rkt.1_intron_1_0_chr9_106792634_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| AAGAACTTATAAAGTTTTGCCGTGAAGCAAGAGGCAAAGAGAATCGGTTGGTAAGTAGCTGGTGACCCACCCAGCCTGTCCCTGGCTCTTGGGCACTGTCCACTTTGTGATGGGACTTGTATATATACTAATGTGAGGTTTAAGTGTTTTTATCCTCTATTTATTAAACATTGCTATAAGCTGAATATTGGTAAGACCAGGAGAGAAAATGACAGGCAACTGCAAGGCAGTGATGCCAGATGCCATGAAG .........................................................(((((........)))))...(((((((.((....(((..((((...........)))).))).....(((((((((..(((((((((((((................)))))))....))))))..))))))))).))))))).)).............................................. ..................................................51..........................................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTAGCTGGTGACCCACCCAGCCTGTCCCTGGCTCTTGGGCACTGTCCA.................................................................................................................................................... | 52 | 1 | 61.00 | 61.00 | 61.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAAGCAAGAGGCAAAGAGAATCGGT.......................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ......................TGAAGCAAGAGGCAAAGAGAATCGGTT......................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...................CCGTGAAGCAAGAGGCAAAGAGAATC............................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TTGGTAAGACCAGGAGAGAAAATGACA.................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................TAAGACCAGGAGAGAAAATGACAGGC................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................GAAGCAAGAGGCAAAGAGAATCGGTT......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................TGAAGCAAGAGGCAAAGAGAATCGGg.......................................................................................................................................................................................................... | 26 | g | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAGCAAGAGGCAAAGAGAATCGGTTGtgt..................................................................................................................................................................................................... | 29 | tgt | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......TATAAAGTTTTGCCGTGAAGCAAGAGGCAAAG................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAAGCTGAATATTGGTAAGACCAGGAG............................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTGAAGCAAGAGGCAAAGAGAA............................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................GAAGCAAGAGGCAAAGAGAATCGGTTG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....CTTATAAAGTTTTGCCGTGAAGCAAGA.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................GCCGTGAAGCAAGAGGCAAAGAGAATCt............................................................................................................................................................................................................ | 28 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................AGCAAGAGGCAAAGAGAATCGGTTGtg...................................................................................................................................................................................................... | 27 | tg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTGAAGCAAGAGGCAAAGAGAATCGGTT......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................AAGCAAGAGGCAAAGAGAATCGGTTG........................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| AAGAACTTATAAAGTTTTGCCGTGAAGCAAGAGGCAAAGAGAATCGGTTGGTAAGTAGCTGGTGACCCACCCAGCCTGTCCCTGGCTCTTGGGCACTGTCCACTTTGTGATGGGACTTGTATATATACTAATGTGAGGTTTAAGTGTTTTTATCCTCTATTTATTAAACATTGCTATAAGCTGAATATTGGTAAGACCAGGAGAGAAAATGACAGGCAACTGCAAGGCAGTGATGCCAGATGCCATGAAG .........................................................(((((........)))))...(((((((.((....(((..((((...........)))).))).....(((((((((..(((((((((((((................)))))))....))))))..))))))))).))))))).)).............................................. ..................................................51..........................................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................AATATTGGTAAGACaatt..................................................... | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................CCTCTATTTATTAActa................................................................................... | 17 | cta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |