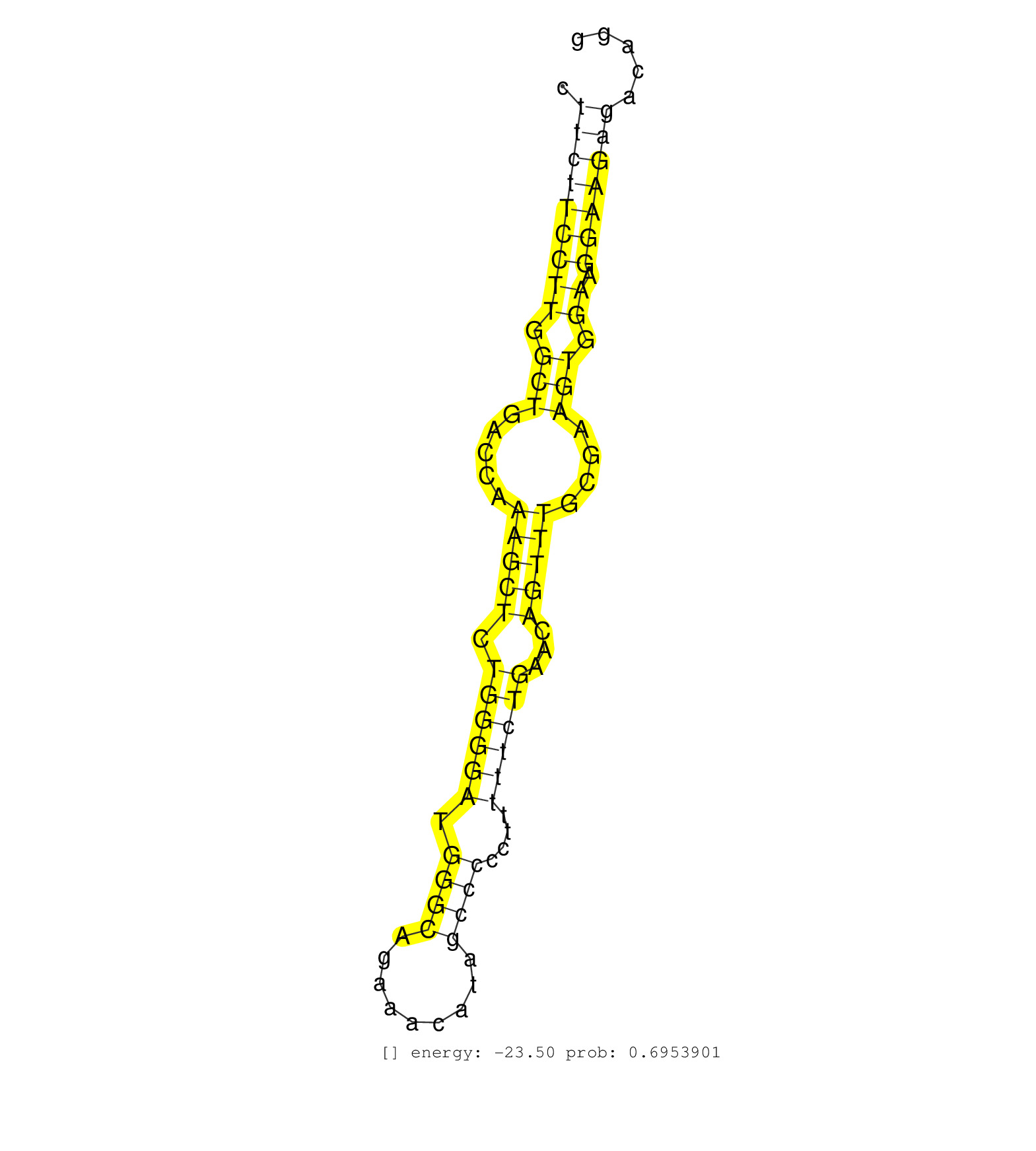
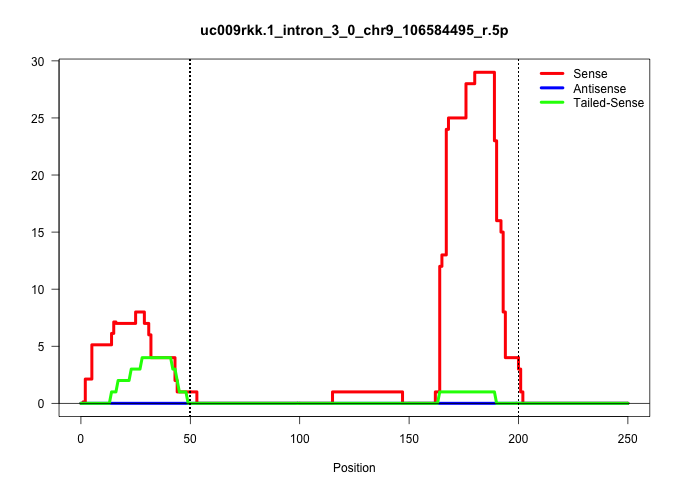
| Gene: Tex264 | ID: uc009rkk.1_intron_3_0_chr9_106584495_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| CCTGCTGGACTGGATGCTATGAGTTGCTAAAAGTGAGCCTGACTTTTTCGGTGAGCCTTCCCATCCCCTGTCCTCGCCTTTCTCTATTGGGTGTGGGGAAATTTGCCTTACTTCTTCCTTGGCTGACCAAAGCTCTGGGGATGGGCAGAAACATAGCCCCCTTTTTCTGAACAGTTTGCGAAGTGGAAGGAAGAGACAGGTTGGGAGCTAAGTTCCCTTAAACAGAAGTGTGTGGCAGTGCTTGTCACTC ...............................................................................................................(((((((((.(((.....(((((.((((((.((((.........))))....))))))...)))))....))).)).)))))))....................................................... ..............................................................................................................111......................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TTCTGAACAGTTTGCGAAGTGGAAGG............................................................ | 26 | 1 | 16.00 | 16.00 | 8.00 | - | - | 3.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTCTGAACAGTTTGCGAAGTGGAAG............................................................. | 25 | 1 | 12.00 | 12.00 | 6.00 | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| .......................................................................................................................................................................TGAACAGTTTGCGAAGTGGAAGGAAG......................................................... | 26 | 1 | 8.00 | 8.00 | - | - | 1.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................TGCGAAGTGGAAGGAAGAGACAGGTTGGG............................................. | 29 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCTGAACAGTTTGCGAAGTGGAAGG............................................................ | 25 | 1 | 5.00 | 5.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAACAGTTTGCGAAGTGGAAGGAAGA........................................................ | 27 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .CTGCTGGACTGGATGCTATGAGTTGCTAAAA.......................................................................................................................................................................................................................... | 31 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGCTGACCAAAGCTCTGGGGATGGGt........................................................................................................ | 27 | t | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGCTGGACTGGATGCTATGAGTTG................................................................................................................................................................................................................................ | 24 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGCTGGACTGGATGCTATGAGTTGC............................................................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGCTGGACTGGATGCTATGAGTTGCTA............................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGCGAAGTGGAAGGAAGAGACAGGT................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGACTGGATGCTATGAGTTGCTAAAA.......................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGCTGGACTGGATGCTATGAGTTGt............................................................................................................................................................................................................................... | 25 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCTGAACAGTTTGCGAAGTGGAAGGAA.......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGGACTGGATGCTATGAGTTGC............................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...GCTGGACTGGATGCTA....................................................................................................................................................................................................................................... | 16 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TAAAAGTGAGCCTGACTTTTTCGGTG..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TTGCTAAAAGTGAGCCTGAtt.............................................................................................................................................................................................................. | 21 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....TGGACTGGATGCTATGAGTTGCTAAA........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TTTTCTGAACAGTTTGCGAAGTGGAAG............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................................................TGCGAAGTGGAAGGAAGAGACAGGTT................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCTAAAAGTGAGCCTGACT.............................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............GCTATGAGTTGCTAAAAGTGAGCCTGAC............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTTTTCTGAACAGTTTGCGAAGTGG................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TGAACAGTTTGCGAAGTGGAAG............................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGCTATGAGTTGCTAAAAGTGAGCCTGAC............................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TCCTTGGCTGACCAAAGCTCTGGGGATGGGCA....................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTCTGAACAGTTTGCGAAGTGGAAtg............................................................ | 26 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................TATGAGTTGCTAAAAGTGAGCCTGtctt............................................................................................................................................................................................................. | 28 | tctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...GCTGGACTGGATGCTATG..................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................AAGTGGAAGGAAGAGACAGG.................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GAACAGTTTGCGAAGTGGAAGGAAGA........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................AAAAGTGAGCCTGACTTactt......................................................................................................................................................................................................... | 21 | actt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................TCTGAACAGTTTGCGAAGTGGAAGGA........................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCTGAACAGTTTGCGAAGTGGAAG............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGCTATGAGTTGCTAAAAGTGAGCCTGt................................................................................................................................................................................................................ | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTTTTCTGAACAGTTTGCGAAGTGGAA.............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CTGCTGGACTGGATG.......................................................................................................................................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTGCTGGACTGGATGCTATGAGTTGCTAAAAGTGAGCCTGACTTTTTCGGTGAGCCTTCCCATCCCCTGTCCTCGCCTTTCTCTATTGGGTGTGGGGAAATTTGCCTTACTTCTTCCTTGGCTGACCAAAGCTCTGGGGATGGGCAGAAACATAGCCCCCTTTTTCTGAACAGTTTGCGAAGTGGAAGGAAGAGACAGGTTGGGAGCTAAGTTCCCTTAAACAGAAGTGTGTGGCAGTGCTTGTCACTC ...............................................................................................................(((((((((.(((.....(((((.((((((.((((.........))))....))))))...)))))....))).)).)))))))....................................................... ..............................................................................................................111......................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT1() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|