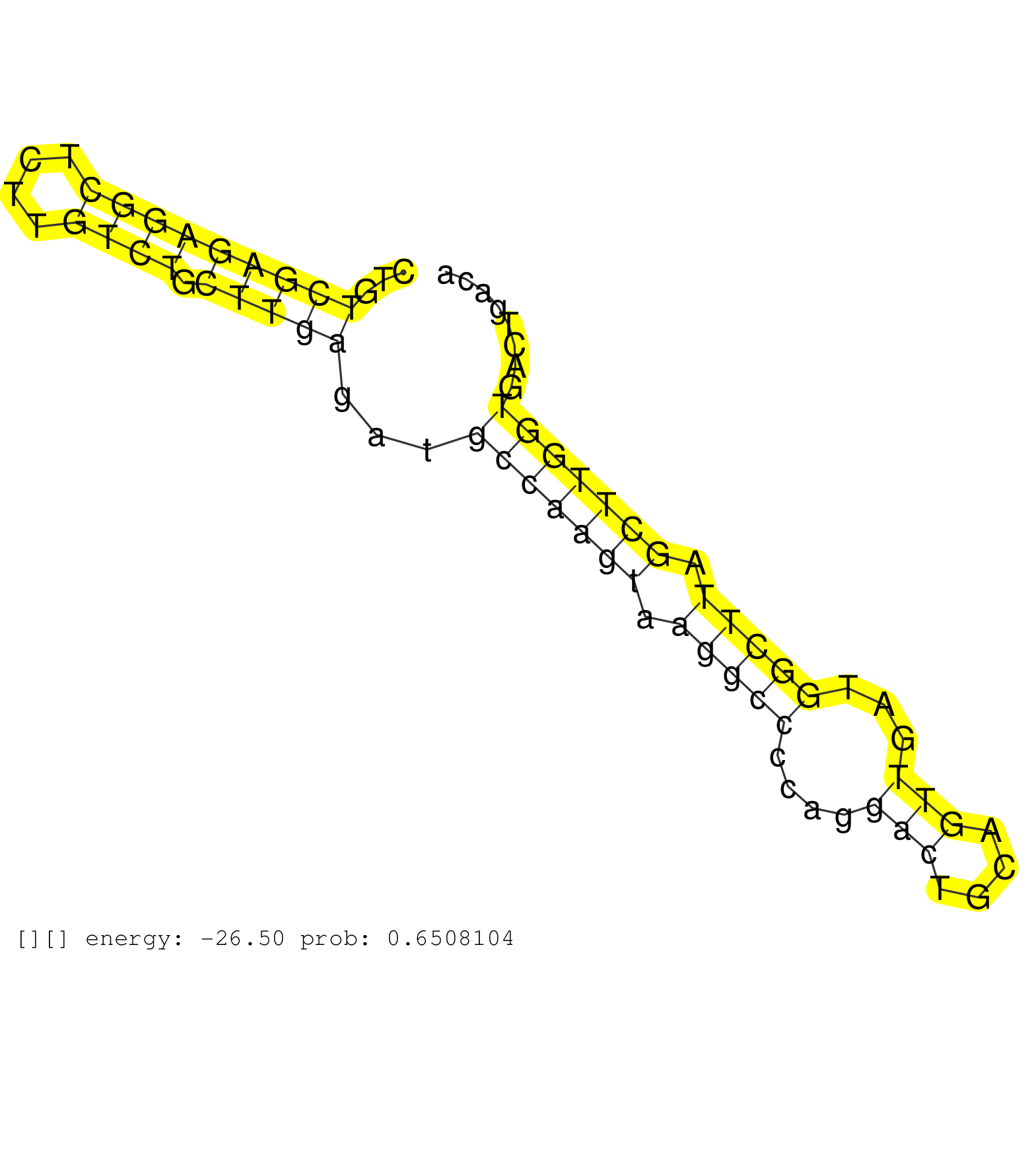
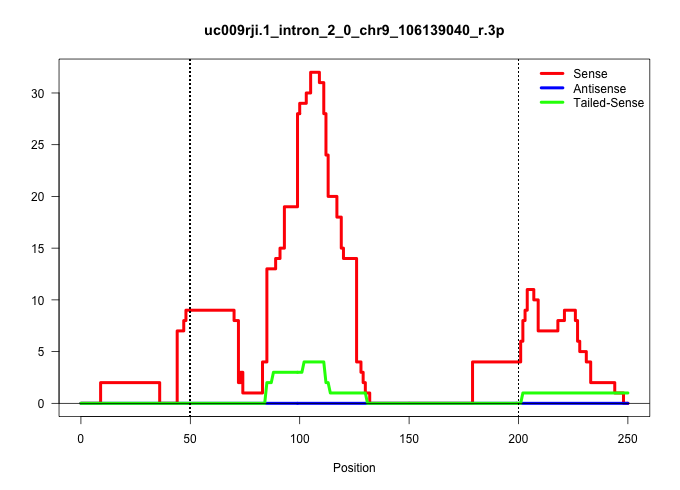
| Gene: Alas1 | ID: uc009rji.1_intron_2_0_chr9_106139040_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| AGTCAGCCCTAACAGGTTCTGGGTGTTTGTACTCATCCAGCAGATGCTAACTGTCGAGAGGCTCTTGTCTGCTTGAGATGCCAAGTAAGGCCCCAGGACTGCAGTTGATGGCTTAGCTTGGTGACTGACACAGAGTCAGTTCCTGGAGGCTGTGCGGTGCTGCTCATGAGGGCCCAGCCTAACAGGACTTCTCCTCCCAGGTAAAGCGTTCGGCTGTGTTGGAGGATACATTGCCAGCACGAGTTTGCTG .....................................................(((((((((....)))).)))))...(((((((.(((((....(((....)))...))))).)))))))................................................................................................................................ ..................................................51.............................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TGCAGTTGATGGCTTAGCTTGGTGACT............................................................................................................................ | 27 | 1 | 9.00 | 9.00 | 4.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TGCTAACTGTCGAGAGGCTCTTGTCTGC.................................................................................................................................................................................. | 28 | 1 | 6.00 | 6.00 | - | 1.00 | - | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................TAAGGCCCCAGGACTGCAGTTGATGGCT......................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................AGCGTTCGGCTGTGTTGGAGGATA...................... | 24 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGGACTGCAGTTGATGGCTTAGCTT................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TAACAGGACTTCTCCTCCCAGGTAAAGCGT......................................... | 30 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TAAGGCCCCAGGACTGCAGTTGATGG........................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TAACAGGTTCTGGGTGTTTGTACTCAT...................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TAAGGCCCCAGGACTGCAGTTGATGGt.......................................................................................................................................... | 27 | t | 2.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................AGTAAGGCCCCAGGACTGCAGTTGATGGC.......................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TAAGGCCCCAGGACTGCAGTTGATGGC.......................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AAAGCGTTCGGCTGTGTTGGAGGATACATTG................. | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTTGATGGCTTAGCTTGGTGACTGACAt....................................................................................................................... | 29 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGATGGCTTAGCTTGGTGACTGACA........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................AGTAAGGCCCCAGGACTGCAGTTGAT............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGATGCCAAGTAAGGCCCCAGGACTGC.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TAAAGCGTTCGGCTGTGTTGGAGGAT....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGCGTTCGGCTGTGTTGGAGGATACAT................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................GCAGTTGATGGCTTAGCTTGGTGACT............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGATGGCTTAGCTTGGTGACTGACACA...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GTTGATGGCTTAGCTTGGTGACTGAC......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AAAGCGTTCGGCTGTGTTGGAGGATAa..................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAACTGTCGAGAGGCTCTTGTCTGCTT................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................GAGGATACATTGCCAGCACGAGTTTGC.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................GCCCCAGGACTGCAGTTGATGGCTTAGC..................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AACTGTCGAGAGGCTCTTGTCTGCTT................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TAAAGCGTTCGGCTGTGTTGGAGGA........................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TAACAGGACTTCTCCTCCCAGGTAAAGC........................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................................AAGCGTTCGGCTGTGTTGGAGGATACATTG................. | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AAAGCGTTCGGCTGTGTTGGAGGAT....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................TGCTAACTGTCGAGAGGCTCTTGTCT.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................AGTTGATGGCTTAGCTTGGTGACTGA.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CCCAGGACTGCAGTTGATGGCTTAGC..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................CAGGACTGCAGTTGATGGCT......................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................ATTGCCAGCACGAGTTTGCTGatt | 24 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................TTGGAGGATACATTGCCAGCACGAGT...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................CAGGACTGCAGTTGATGGCTTAGCTTG.................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGCCCCAGGACTGCAGTTGATGGCTa........................................................................................................................................ | 26 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| AGTCAGCCCTAACAGGTTCTGGGTGTTTGTACTCATCCAGCAGATGCTAACTGTCGAGAGGCTCTTGTCTGCTTGAGATGCCAAGTAAGGCCCCAGGACTGCAGTTGATGGCTTAGCTTGGTGACTGACACAGAGTCAGTTCCTGGAGGCTGTGCGGTGCTGCTCATGAGGGCCCAGCCTAACAGGACTTCTCCTCCCAGGTAAAGCGTTCGGCTGTGTTGGAGGATACATTGCCAGCACGAGTTTGCTG .....................................................(((((((((....)))).)))))...(((((((.(((((....(((....)))...))))).)))))))................................................................................................................................ ..................................................51.............................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|