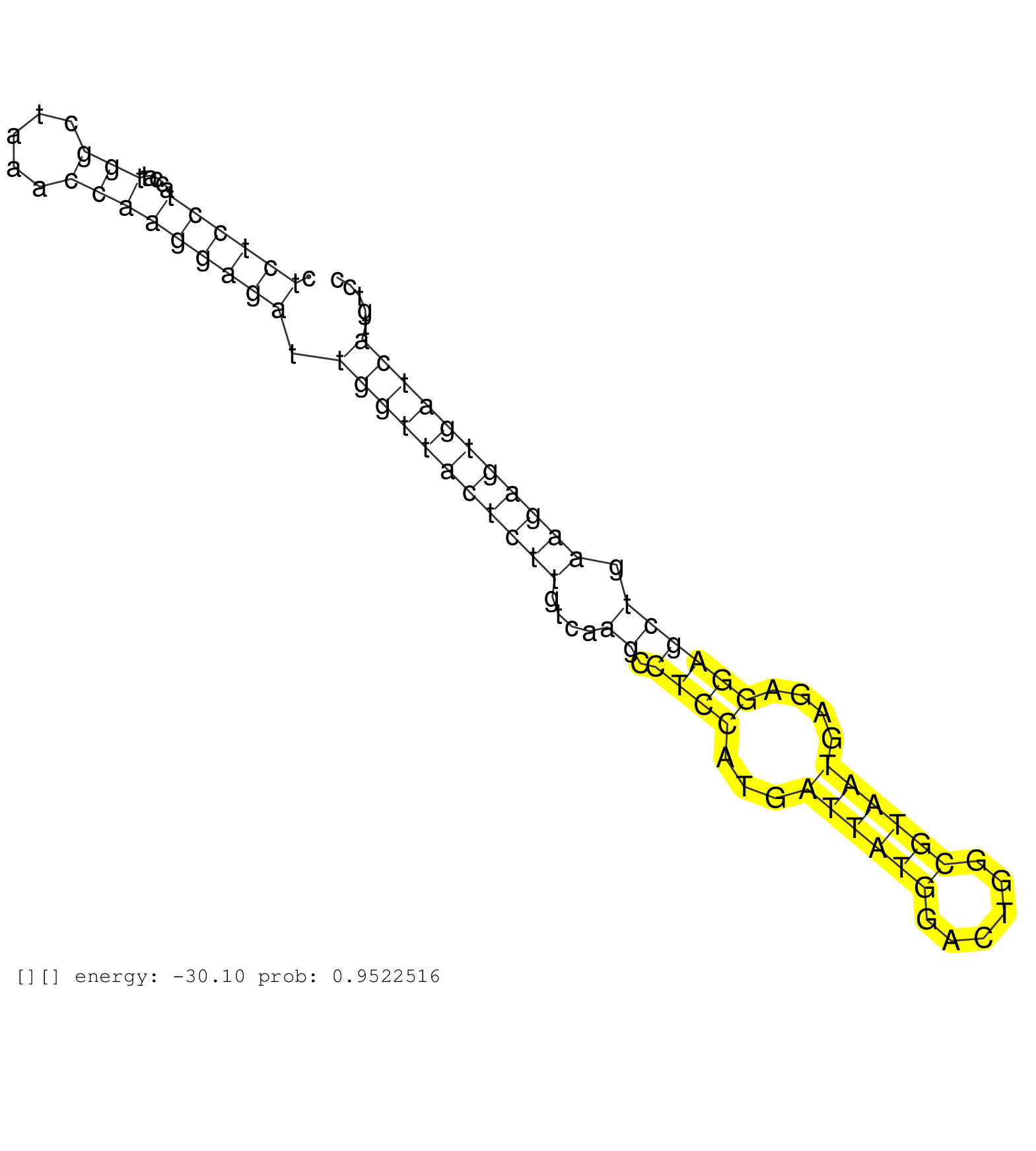

| Gene: 1110001D15Rik | ID: uc009rix.1_intron_24_0_chr9_105954036_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.mut |
(18) TESTES |
| AGAGGGGTTTGGACGGGCTCCCTGGCCATCCTGGTGAAGAGGGAGACCACGTGAGTACTGCATCTTTTCTCGTTCTCAGCCTCCCTTCCTTTCTTGTCCCCTCTCCTACCATTGGCTAAACCAAGGAGATTGGTTACTCTTGTCAAGCCTCCATGATTATGGACTGGCGTAATGAGAGGAGCTGAAGAGTGATCATGTCCATTCCTAATATATAACACAGTCCCTCAGTTCTTTCTTGAGTGGCAGTGAA .....................................................................................................((((((.....(((.....))))))))).(((((((((((....((.((((...((((((......))))))....)))))).)))))))))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................CCTCCATGATTATGGACTGGCGTAATGAGAGGA...................................................................... | 33 | 1 | 129.00 | 129.00 | 45.00 | 33.00 | 23.00 | 6.00 | 7.00 | - | 3.00 | 3.00 | 2.00 | 3.00 | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAGAGGA...................................................................... | 32 | 1 | 36.00 | 36.00 | 4.00 | 12.00 | 14.00 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CCTCCATGATTATGGACTGGCGTAATGAGAGG....................................................................... | 32 | 1 | 20.00 | 20.00 | 13.00 | - | - | 1.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAGA......................................................................... | 29 | 1 | 5.00 | 5.00 | 2.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TCCATGATTATGGACTGGCGTAATGAGAGGA...................................................................... | 31 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAGAG........................................................................ | 30 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CCTCCATGATTATGGACTGGCGTAATGAGggga...................................................................... | 33 | ggga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................CCATGATTATGGACTGGCGTAATGAGAGGA...................................................................... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GCCTCCATGATTATGGACTGGCGTAATGAcac........................................................................ | 32 | cac | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAtag........................................................................ | 30 | tag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAGAGa....................................................................... | 31 | a | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGATTATGGACTGGCGTAATGAGAGta...................................................................... | 27 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................CTCCATGATTATGGACTGGCGTAATGAGAGG....................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AGAGGGGTTTGGACGGGCTCCCTGGCCATCCTGGTGAAGAGGGAGACCACGTGAGTACTGCATCTTTTCTCGTTCTCAGCCTCCCTTCCTTTCTTGTCCCCTCTCCTACCATTGGCTAAACCAAGGAGATTGGTTACTCTTGTCAAGCCTCCATGATTATGGACTGGCGTAATGAGAGGAGCTGAAGAGTGATCATGTCCATTCCTAATATATAACACAGTCCCTCAGTTCTTTCTTGAGTGGCAGTGAA .....................................................................................................((((((.....(((.....))))))))).(((((((((((....((.((((...((((((......))))))....)))))).)))))))))))....................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|