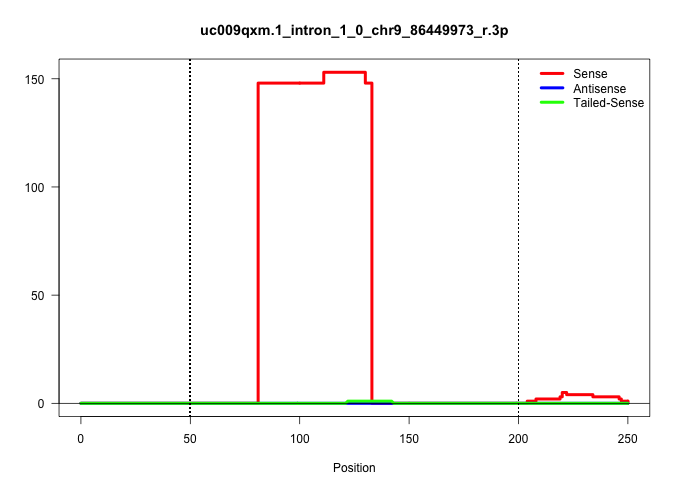
| Gene: Pgm3 | ID: uc009qxm.1_intron_1_0_chr9_86449973_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(9) TESTES |
| CTGTCTTGAAAAATGAAAAATAAAACAGGAAATTTTATTATTGCAGTTTCTTCTCACCTGCTCTGGTGTTTCCCTGCTGCACACTGTAGTGTGGCAACCTGAAACGCCAGTCTGTTACTCTCTGGCAAGACTATTTAGAACAGGTTAAGCTTTTGAAAGTGAGTATAACAGAGTAACACATACAGCATATTTTTGCTCAGGTCGCGGACAGGAGAGTTATTAGCACCACGGATGCTGAGAGACAAGCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................CACTGTAGTGTGGCAACCTGAAACGCCAGTCTGTTACTCTCTGGCAAGACTA..................................................................................................................... | 52 | 1 | 148.00 | 148.00 | 148.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................CTGTTACTCTCTGGCAAGA........................................................................................................................ | 19 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TTAGCACCACGGATGCTGAGAGACAAG.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................CAGGAGAGTTATTAGCACCACGGATG................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................................TAGCACCACGGATGCTGAGAGACAAGC... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................................................................................................................TAGCACCACGGATGCTGAGAGACAAGCAGT | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................TGGCAAGACTATTTAGAACcc........................................................................................................... | 21 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................CGGACAGGAGAGTTATTA............................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| CTGTCTTGAAAAATGAAAAATAAAACAGGAAATTTTATTATTGCAGTTTCTTCTCACCTGCTCTGGTGTTTCCCTGCTGCACACTGTAGTGTGGCAACCTGAAACGCCAGTCTGTTACTCTCTGGCAAGACTATTTAGAACAGGTTAAGCTTTTGAAAGTGAGTATAACAGAGTAACACATACAGCATATTTTTGCTCAGGTCGCGGACAGGAGAGTTATTAGCACCACGGATGCTGAGAGACAAGCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................AGGTTAAGCTTTTGcaga............................................................................................... | 18 | caga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - |