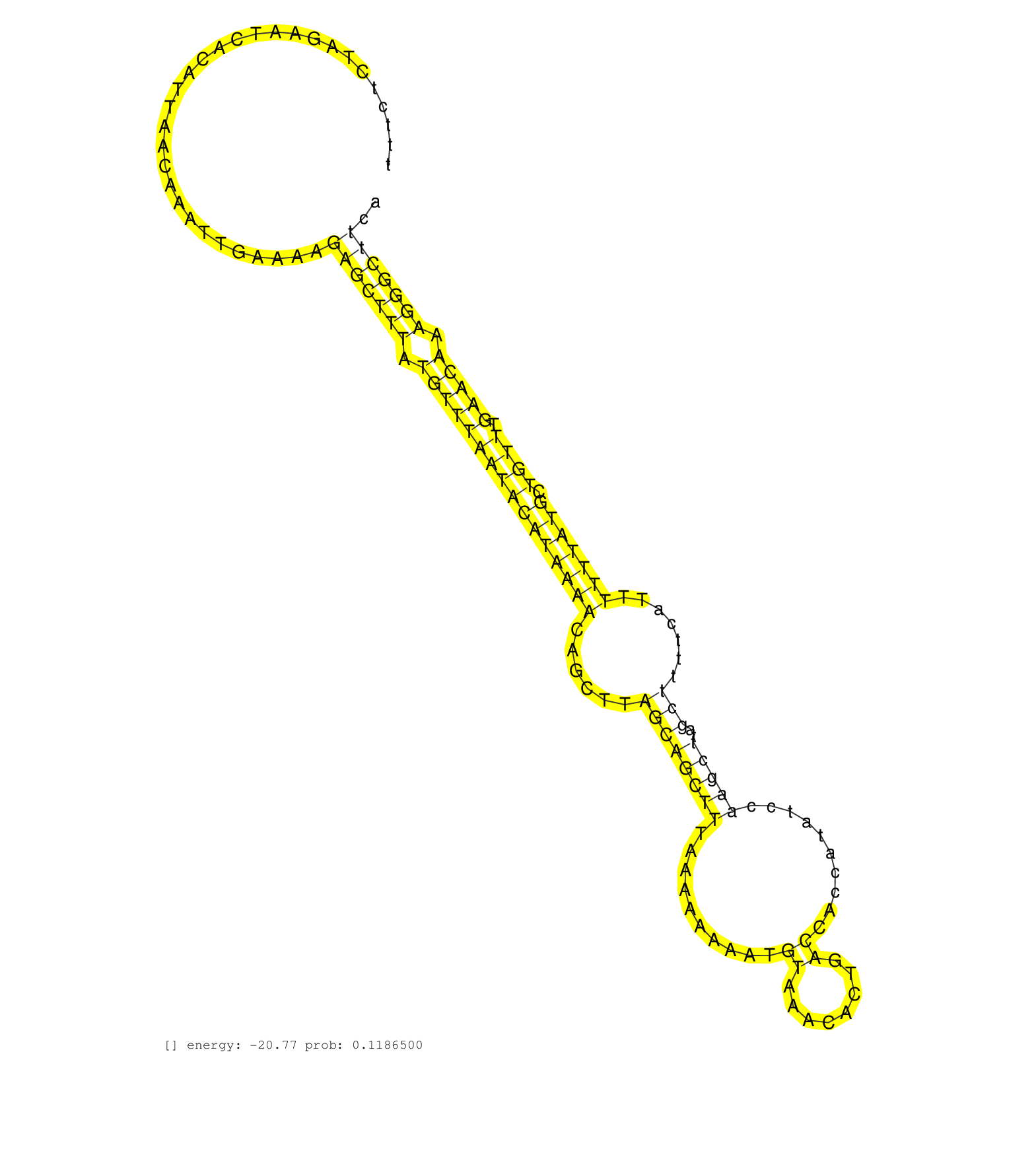

| Gene: AK017040 | ID: uc009qsg.1_intron_1_0_chr9_75259672_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| AAGCGTGCTTAGGTTTGCATGTAGAGTTTCTTTCCTACAGCATAAAAATAATTAAAGCTTCTTTCTCTAGAATCACATTAACAAATTGAAAAGAGCTTTATGTTTAATACATAAAACAGCTTAGCAGCTTTAAAAAAAATGTAAACACTGACCACCATATCCAAGCTTAGCTTTTCATTTTTTATGCTGTTTGAACAAAGGGCTTCAGAAATTCACCAGCATTGCTAAGAAATCGTCTCCCCAGCCCCCA ............................................................................................(((((((.((((((((((((((((......((((((((..........((........))..........)))))..))).......))))))).)))).))))).)))))))............................................. .............................................................62...............................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................CTAGAATCACATTAACAAATTGAAAAGAGCTTTATGTTTAATACATAAAACA.................................................................................................................................... | 52 | 1 | 63.00 | 63.00 | 63.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGCTGTTTGAACAAAGGGCTTCAGAAA....................................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................TTTTTTATGCTGTTTGAACAAAGGGC............................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................TTCACCAGCATTGCTAAGAAA.................. | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TCTAGAATCACATTAACAAATTGAAAAGAGC.......................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................TAGAATCACATTAACAAATTGAAAAGAGC.......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................TGAACAAAGGGCTTCAGAAATTCACCAGCAT............................ | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AATGTAAACACTGACCACCATATCCAAGCTTA................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGAAATTCACCAGCATTGCTAAGAAAc................. | 28 | c | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGCTAAGAAATCGTCTCCCCAGCCCCC. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TTCACCAGCATTGCTAAGAAATCGTCTCCC......... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TAAGAAATCGTCTCCCCAGCCCCC. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................CAGCATTGCTAAGAAATCGTCTCCCCAGt..... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................AGGGCTTCAGAAATTCACCAGCATTGCTAAG..................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................TGAACAAAGGGCTTCAGAAATTCACCAG............................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| AAGCGTGCTTAGGTTTGCATGTAGAGTTTCTTTCCTACAGCATAAAAATAATTAAAGCTTCTTTCTCTAGAATCACATTAACAAATTGAAAAGAGCTTTATGTTTAATACATAAAACAGCTTAGCAGCTTTAAAAAAAATGTAAACACTGACCACCATATCCAAGCTTAGCTTTTCATTTTTTATGCTGTTTGAACAAAGGGCTTCAGAAATTCACCAGCATTGCTAAGAAATCGTCTCCCCAGCCCCCA ............................................................................................(((((((.((((((((((((((((......((((((((..........((........))..........)))))..))).......))))))).)))).))))).)))))))............................................. .............................................................62...............................................................................................................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................TTTCCTACAGCATAAAagtc............................................................................................................................................................................................................ | 20 | agtc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................CACTGACCACCATAgggg........................................................................................... | 18 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |