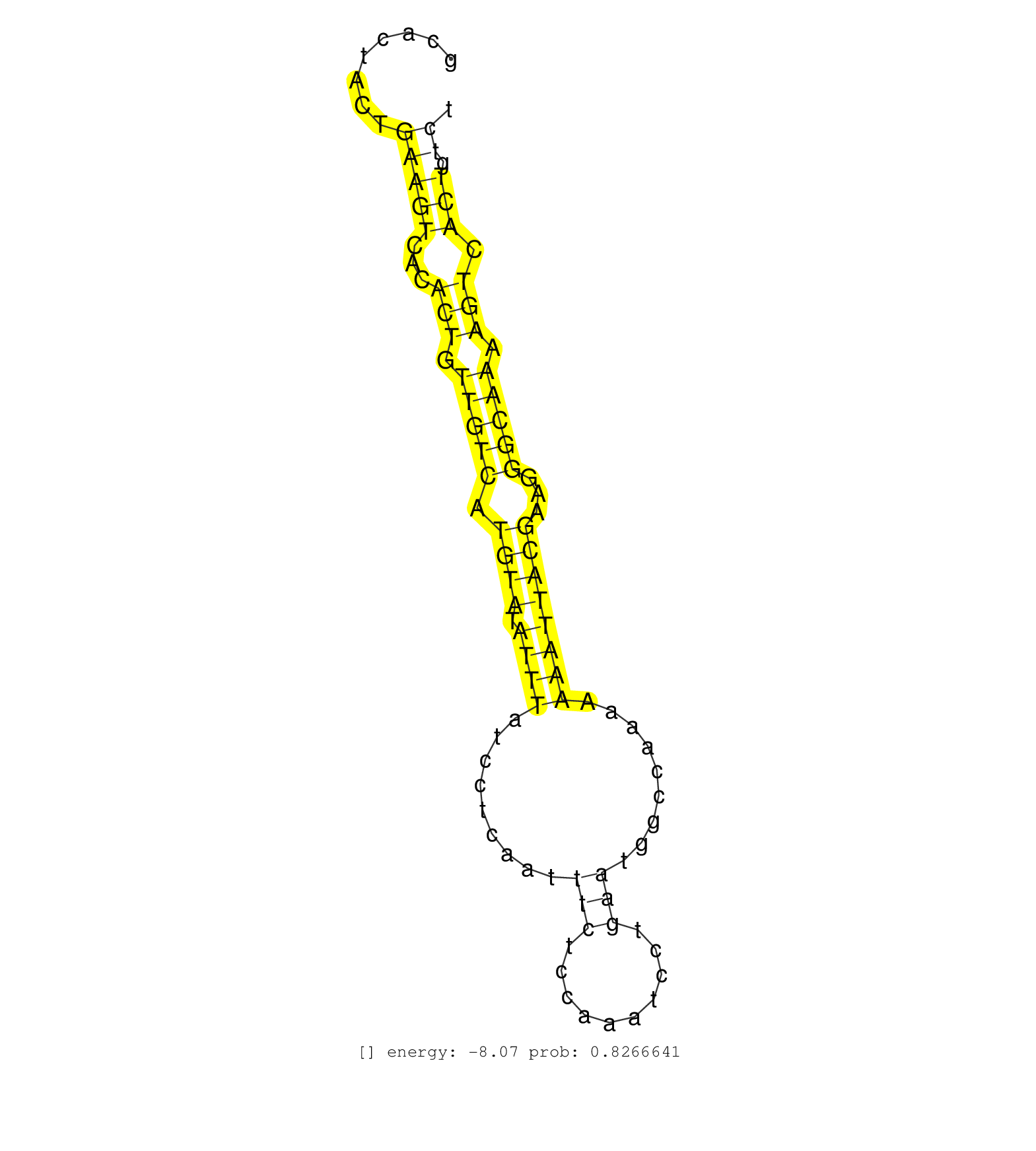

| Gene: Rab27a | ID: uc009qqv.1_intron_4_0_chr9_72935008_f.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(23) TESTES |
| TCATTTTTTTTCATGTAATTTGGCACACTGAATTAAACTGAAACACTGTCGCCTCCTCTCTGGGCACTACTGAAGTCACACTGTTGTCATGTATATTTATCCTCAATTTCTCCAAATCCTGAATGGCCAAAAAAATTACGAAGGGCAAAAGTCACTGTCTTTCTTCCCACATTGTTACCCCAGTGATTTCTGTCTTCCAGAATCCCCTATTTTGAAACCAGTGCTGCCAACGGGACAAACATAAGCCACG .......................................................................(((((...(((.(((((.((((.((((.........(((..........))).........))))))))...))))).))).))).))........................................................................................... ...............................................................64..............................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................ACTGAAGTCACACTGTTGTCATGTATATTT........................................................................................................................................................ | 30 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................ACTGAAGTCACACTGTTGTCATGTATATT......................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGGGCACTACTGAAGTCACACTGTTGTC.................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGAAACACTGTCGCCTCCTCTCT............................................................................................................................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAATGGCCAAAAAAATTACGAAGGGCA....................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AAAATTACGAAGGGCAAAAGTCAtt.............................................................................................. | 25 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TCTCTGGGCACTACTGAAGTCACACTG....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................GAATGGCCAAAAAAATTACGAAGGGC........................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GAAACCAGTGCTGCCAACGGGA............... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAATGGCCAAAAAAATTACGAAGGG......................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGAAACCAGTGCTGCCAACGGGACAAA........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGGGCACTACTGAAGTCACACTGTTGTga................................................................................................................................................................. | 29 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TGGCACACTGAATTAAACTGAAACACTGTC........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TCCTCAATTTCTCCAAATCCTGAATGGCCA......................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TATTTATCCTCAATTTCTCCAAATCC................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGAAACACTGTCGCCTCCTCTCTGGGa......................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................TCCTGAATGGCCAAAAAAATTACGAAGtgc........................................................................................................ | 30 | tgc | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAATGGCCAAAAAAATTACGAAGGGCAA...................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTACGAAGGGCAAAAGTCACTGTCTTT........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCCTGAATGGCCAAAAAAATTACGAAG........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................TGAATTAAACTGAAACACTGTCGCCTCCTC................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGAATGGCCAAAAAAATTACGAAGGGC........................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGAAGTCACACTGTTGTCATGTATATT......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................AAAAAAATTACGAAGGGCAAAAGTCAC............................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GAATGGCCAAAAAAATTACGAAGGGCAAA..................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGAAGTCACACTGTTGTCATGTATATTTA....................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TACTGAAGTCACACTGTTGTCATGTATAT.......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TCCTCTCTGGGCACTACTGAAGTCAC........................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................TGAAGTCACACTGTTGTCATGTATATTTATt..................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGGGCACTACTGAAGTCACACTGTTGTCATG............................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................CTCCTCTCTGGGCACTACTGAAGTCACAC......................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................CTCCTCTCTGGGCACTACTGAAGTCACACT........................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTTGAAACCAGTGCTGCCAACGGG................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................CCACATTGTTACCCCAGTGAT............................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TTAAACTGAAACACTGTCGCCTCCTCTCT............................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TATTTTGAAACCAGTGCTGCCAACGGGA............... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................AATTAAACTGAAACACTGTCGCCTC................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCATTTTTTTTCATGTAATTTGGCACACTGAATTAAACTGAAACACTGTCGCCTCCTCTCTGGGCACTACTGAAGTCACACTGTTGTCATGTATATTTATCCTCAATTTCTCCAAATCCTGAATGGCCAAAAAAATTACGAAGGGCAAAAGTCACTGTCTTTCTTCCCACATTGTTACCCCAGTGATTTCTGTCTTCCAGAATCCCCTATTTTGAAACCAGTGCTGCCAACGGGACAAACATAAGCCACG .......................................................................(((((...(((.(((((.((((.((((.........(((..........))).........))))))))...))))).))).))).))........................................................................................... ...............................................................64..............................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................GTTACCCCAGTGATTTCTGTCTTCCA................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 |
| ..................TTTGGCACACTGAATTAAACTGAAACACTGT......................................................................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GCACTACTGAAGTCACACTGTTGTCATGTA............................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TACTGAAGTCACACTGTTGTCATGTA............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................CTGAATGGCCAAAAAAATT................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................AAGTCACACTGTTGTCATGTATATTTATCCT................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................ATGTATATTTATCCTCAATTTCTCCAAATCCT.................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................CTGTTGTCATGTATATTTATCCTCAATTTCTC.......................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................GCCTCCTCTCTGGGCattt......................................................................................................................................................................................... | 19 | attt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................CCTCTCTGGGCACTACTGAAGTCACA.......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CCAGTGATTTCTGTCTTCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GGGCAAAAGTCACTGTCTTTCTTCCCACA............................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................ATATTTATCCTCAATTTCTCCAAATCCTGAA............................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................GCCTCCTCTCTGGGCacct......................................................................................................................................................................................... | 19 | acct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |