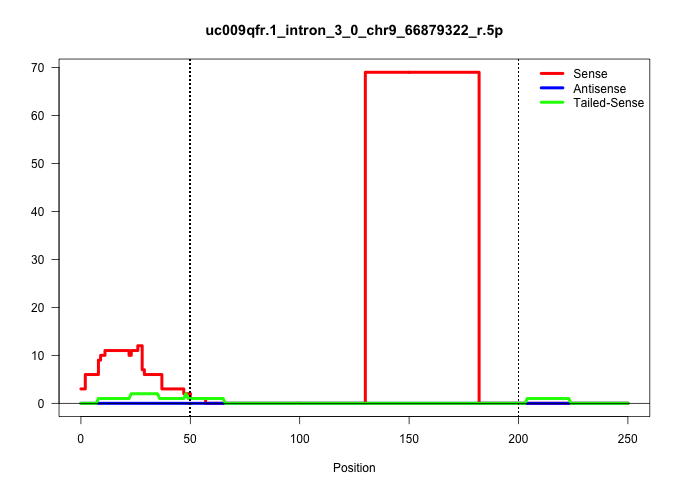
| Gene: Tpm1 | ID: uc009qfr.1_intron_3_0_chr9_66879322_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| TAAGAATAATGGATCAGACCTTGAAAGCATTAATGGCTGCAGAGGATAAGGTACTGATAGCTCACGTATAGTTTTTAGGTTTAACTGCATCCCAGACAGCTTTGAGCTTCCAATGCCTACTGGTCGGTTTGGTGTAATGACTGTACCTCCACTACACCCTCTGCTACTTATATCTTGCTTCAAGTGCTTTCTCTTGGGCCTTTATGTTCTTTGTTTTTCCCCTTCATCGATACTCCGAGGGGTGGCCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................GGTGTAATGACTGTACCTCCACTACACCCTCTGCTACTTATATCTTGCTTCA.................................................................... | 52 | 1 | 69.00 | 69.00 | 69.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATGGATCAGACCTTGAAAGCATTAATGGC..................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..AGAATAATGGATCAGACCTTGAAAGC.............................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AGAATAATGGATCAGACCTT.................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........ATGGATCAGACCTTGAAAGCA............................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........GATCAGACCTTGAAAGCATTAATGGC..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGTTCTTTGTTTTTCCacag.......................... | 20 | acag | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........TGGATCAGACCTTGAAAGCATTAATGGC..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................AAAGCATTAATGGCTGCAGAGGATAt......................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................AGGTACTGATAGCTgagt........................................................................................................................................................................................ | 18 | gagt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................AAAGCATTAATGGCTGCAGAGGATAAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGCAGAGGATAAGGTACTGA................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........ATGGATCAGACCTTGAAAGCATTAATGt...................................................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................GCATTAATGGCTGCAGAGGAT........................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TAAGAATAATGGATCAGACCTTGAAAGCATTAATGGCTGCAGAGGATAAGGTACTGATAGCTCACGTATAGTTTTTAGGTTTAACTGCATCCCAGACAGCTTTGAGCTTCCAATGCCTACTGGTCGGTTTGGTGTAATGACTGTACCTCCACTACACCCTCTGCTACTTATATCTTGCTTCAAGTGCTTTCTCTTGGGCCTTTATGTTCTTTGTTTTTCCCCTTCATCGATACTCCGAGGGGTGGCCAGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................CATCCCAGACAGCTTTGAtgtg................................................................................................................................................. | 22 | tgtg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |