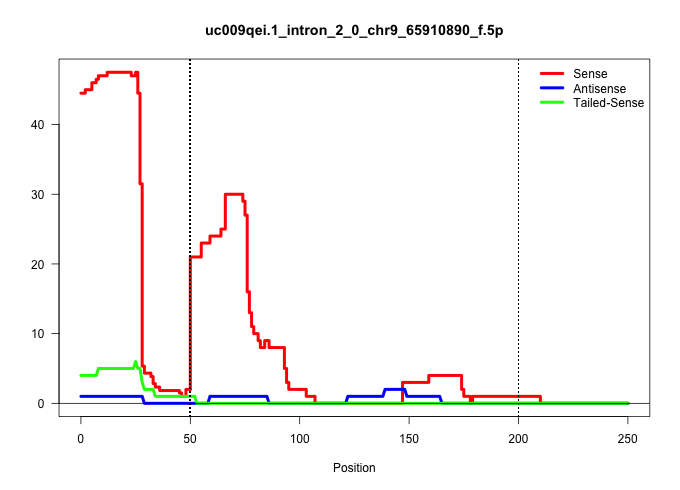
| Gene: Ppib | ID: uc009qei.1_intron_2_0_chr9_65910890_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| GGACTTCATGATCCAGGGTGGAGACTTCACCAGGGGAGATGGCACAGGAGGTAACTTCAGGCCCTCTGAGCAGAAGAGTCTTGACACTGTAGTGTCCAAGGTGGGGTGGCCAGGCATCTCCTTTCACCTGCTCTAGGGGCCGGCTTCTCTCCTGCATTGAGACTAGCTGTACCCTTCTTTGAAAGGACAGCCCAGTGCAATTAACAAGTCTGTAGCTTGCACCACACAATCTGTTGCTTGTGTAAAACAC ..................................................(((......((((((..((((((..(((.((((((((....))))).)))..((((((......))))))..)))..))))))..)))))).(((..((((.......))))..)))..))).............................................................................. ..................................................51.............................................................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................AGAGTCTTGACACTGTAGT............................................................................................................................................................. | 19 | 1 | 12.00 | 12.00 | - | - | - | - | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAAG.............................................................................................................................................................................. | 26 | 1 | 11.00 | 11.00 | - | 1.00 | - | 8.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAAGA............................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGAGCAGAAGAGTCTTGACACTGTAGT............................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGAGCAGAAGAGTCTTGACACTGTAGTG............................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAAGAG............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTCCTGCATTGAGACTAGCTGTACCC............................................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAAGAGT........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGAGCAGAAGAGTCTTGACACTGTAGTGT........................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TTCAGGCCCTCTGAGCAGAAGAGTCTT........................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTCACCAGGGGAGATGGCACAGGAGGaa..................................................................................................................................................................................................... | 28 | aa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGCCCTCTGAGCAGAAGAGTCTTGACA.................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................CACTGTAGTGTCCAAGGTG................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTAACTTCAGGCCCTCTGAGCAGAA............................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TTCAGGCCCTCTGAGCAGAAGAGTCT......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAACTTCAGGCCCTCTGAGCAtaag.............................................................................................................................................................................. | 25 | taag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................TCTGAGCAGAAGAGTCTTGACACTGTAGT............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TCTCCTGCATTGAGACTAGCTGTACCCT........................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGA................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AGACTAGCTGTACCCTTCT........................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TTCACCAGGGGAGATGGCACAGGAGG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TAACTTCAGGCCCTCTGAGCAGAAG.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGAAAGGACAGCCCAGTGCAATTAACAAGTC........................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AACTTCAGGCCCTCTGAGCAGAAaact........................................................................................................................................................................... | 27 | aact | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................TTGACACTGTAGTGTCCAAGGTGGGGT............................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAAGAGTCTTG....................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........TGATCCAGGGTGGAGACTTCACCAGt........................................................................................................................................................................................................................ | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................GGAGACTTCACCAGGGGAGATGGCACAG........................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....TCATGATCCAGGGTGGAGACTTCACCAG......................................................................................................................................................................................................................... | 28 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 |
| ...................................................TAACTTCAGGCCCTCTGAGCAGAAGAGTC.......................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAACTTCAGGCCCTCTGAGCAGAA............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............CCAGGGTGGAGACTTCACCAGGGG...................................................................................................................................................................................................................... | 24 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................ACCAGGGGAGATGGCACA............................................................................................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| .....TCATGATCCAGGGTGGAGACTTCACC........................................................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ....................GAGACTTCACCAGGGGAGATGGC............................................................................................................................................................................................................... | 23 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| ..ACTTCATGATCCAGGGTGGAGACTTCACCA.......................................................................................................................................................................................................................... | 30 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ........TGATCCAGGGTGGAGACTTCACCAGG........................................................................................................................................................................................................................ | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......CATGATCCAGGGTGGAGACTTCAC............................................................................................................................................................................................................................ | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .......ATGATCCAGGGTGGAGACTT............................................................................................................................................................................................................................... | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ............................ACCAGGGGAGATGGCAC............................................................................................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ...CTTCATGATCCAGGGT....................................................................................................................................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - |
| GGACTTCATGATCCAGGGTGGAGACTTCACCAGGGGAGATGGCACAGGAGGTAACTTCAGGCCCTCTGAGCAGAAGAGTCTTGACACTGTAGTGTCCAAGGTGGGGTGGCCAGGCATCTCCTTTCACCTGCTCTAGGGGCCGGCTTCTCTCCTGCATTGAGACTAGCTGTACCCTTCTTTGAAAGGACAGCCCAGTGCAATTAACAAGTCTGTAGCTTGCACCACACAATCTGTTGCTTGTGTAAAACAC ..................................................(((......((((((..((((((..(((.((((((((....))))).)))..((((((......))))))..)))..))))))..)))))).(((..((((.......))))..)))..))).............................................................................. ..................................................51.............................................................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................CCGGCTTCTCTCCTGCATTGAGACTA..................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TTCACCTGCTCTAGGGGCCGGCTTCTC..................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GGCCCTCTGAGCAGAAGAGTCTTGACA.................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |