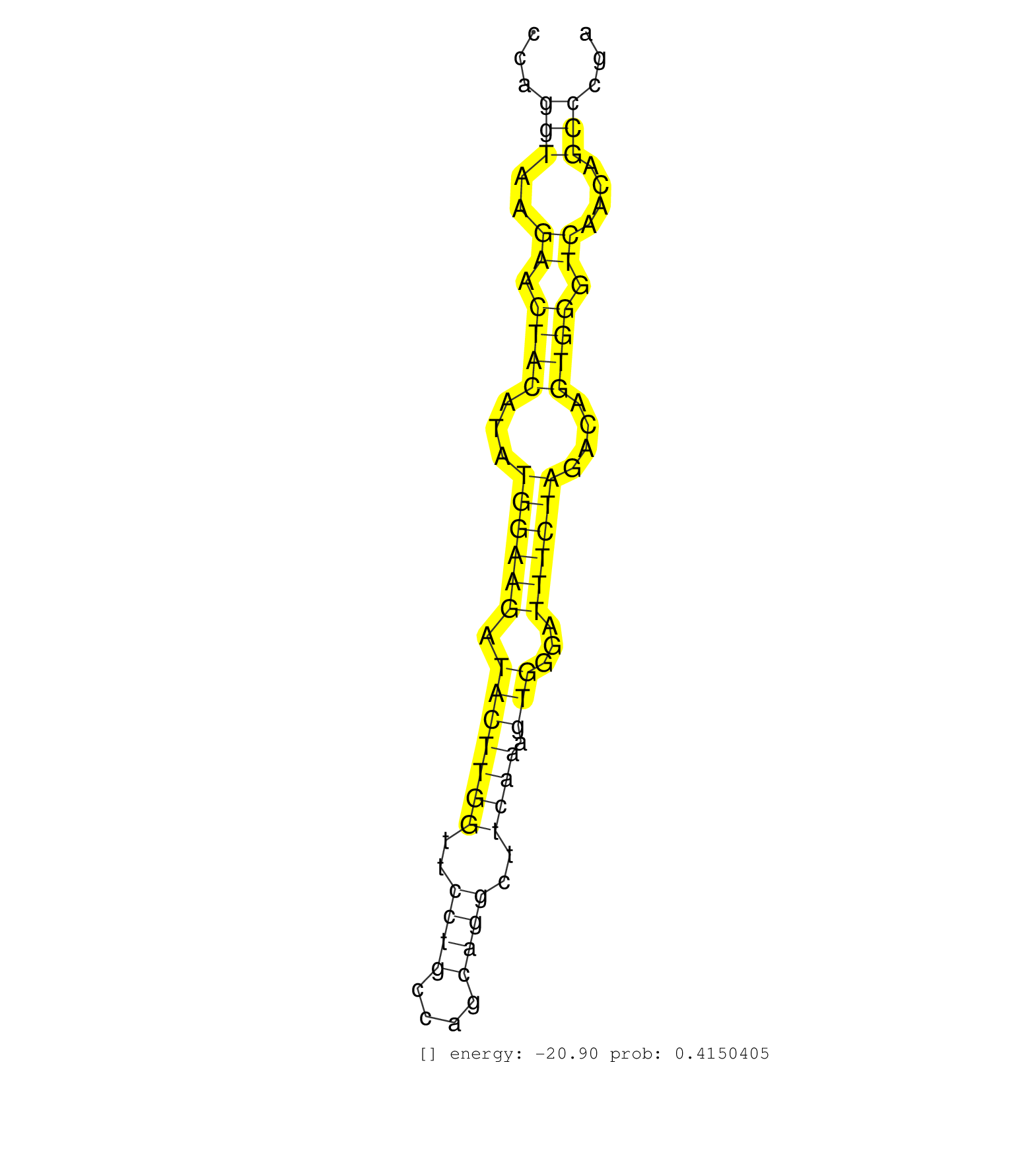

| Gene: Pml | ID: uc009pwn.1_intron_0_0_chr9_58068372_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| AGCCAGCTTCAGGACGGGTGAGCCATCCCCGCCCCCTATCGGAGCCAGGTAAGAACTACATATGGAAGATACTTGGTTCCTGCCAGCAGGCTTCAAAGTGGGATTTCTAGACAGTGGGTCAACAGCCCGATCCCTTGCACTCAACCCAGTGCCTCCTGAACCCTGAACCTCACCGTGACCTTCTTTCCTGCCTAGCCCAGAAAATTAGCCAGCTGGCCGCGGTGAACCGGGAAAGCAAGTTCCGTGTGCT ...............................................(((..((.((((...((((((.(((((((..((((....))))..)))).)))...))))))....)))).))....)))........................................................................................................................... ............................................45...................................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................TAAGAACTACATATGGAAGATACTTGG.............................................................................................................................................................................. | 27 | 1 | 11.00 | 11.00 | 5.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................TGGGATTTCTAGACAGTGGGTCAACAGC............................................................................................................................ | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTCAAAGTGGGATTTCTAGACAGTGG..................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................................TGAACCTCACCGTGACCTTCTTTCCTG............................................................ | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................TAGCCAGCTGGCCGCGG............................ | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTCTAGACAGTGGGTCAA................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CACCGTGACCTTCTTTCCTGCCTAGCCC.................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................TTTCTAGACAGTGGGcat................................................................................................................................. | 18 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................GTAAGAACTACATATGGAAGATACTTGG.............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGGAAGATACTTGGTTCCTGCCAGCAG................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TACATATGGAAGATACTTGGTTCCTGC....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................TGAACCTCACCGTGACCTTCTTTCCTGCC.......................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGGTGAACCGGGAAAGCAAGTT......... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................TCGGAGCCAGGTAAGAACTACATATGGAAGt..................................................................................................................................................................................... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................TGGGATTTCTAGACAGTGGGTCAACAG............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGAAAATTAGCCAGttga.................................. | 18 | ttga | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TTCAAAGTGGGATTTCTAGACAGT....................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TATCGGAGCCAGGTAAGAACT................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCCAGCTTCAGGACGGGTGAGCCATCCCCGCCCCCTATCGGAGCCAGGTAAGAACTACATATGGAAGATACTTGGTTCCTGCCAGCAGGCTTCAAAGTGGGATTTCTAGACAGTGGGTCAACAGCCCGATCCCTTGCACTCAACCCAGTGCCTCCTGAACCCTGAACCTCACCGTGACCTTCTTTCCTGCCTAGCCCAGAAAATTAGCCAGCTGGCCGCGGTGAACCGGGAAAGCAAGTTCCGTGTGCT ...............................................(((..((.((((...((((((.(((((((..((((....))))..)))).)))...))))))....)))).))....)))........................................................................................................................... ............................................45...................................................................................130...................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|