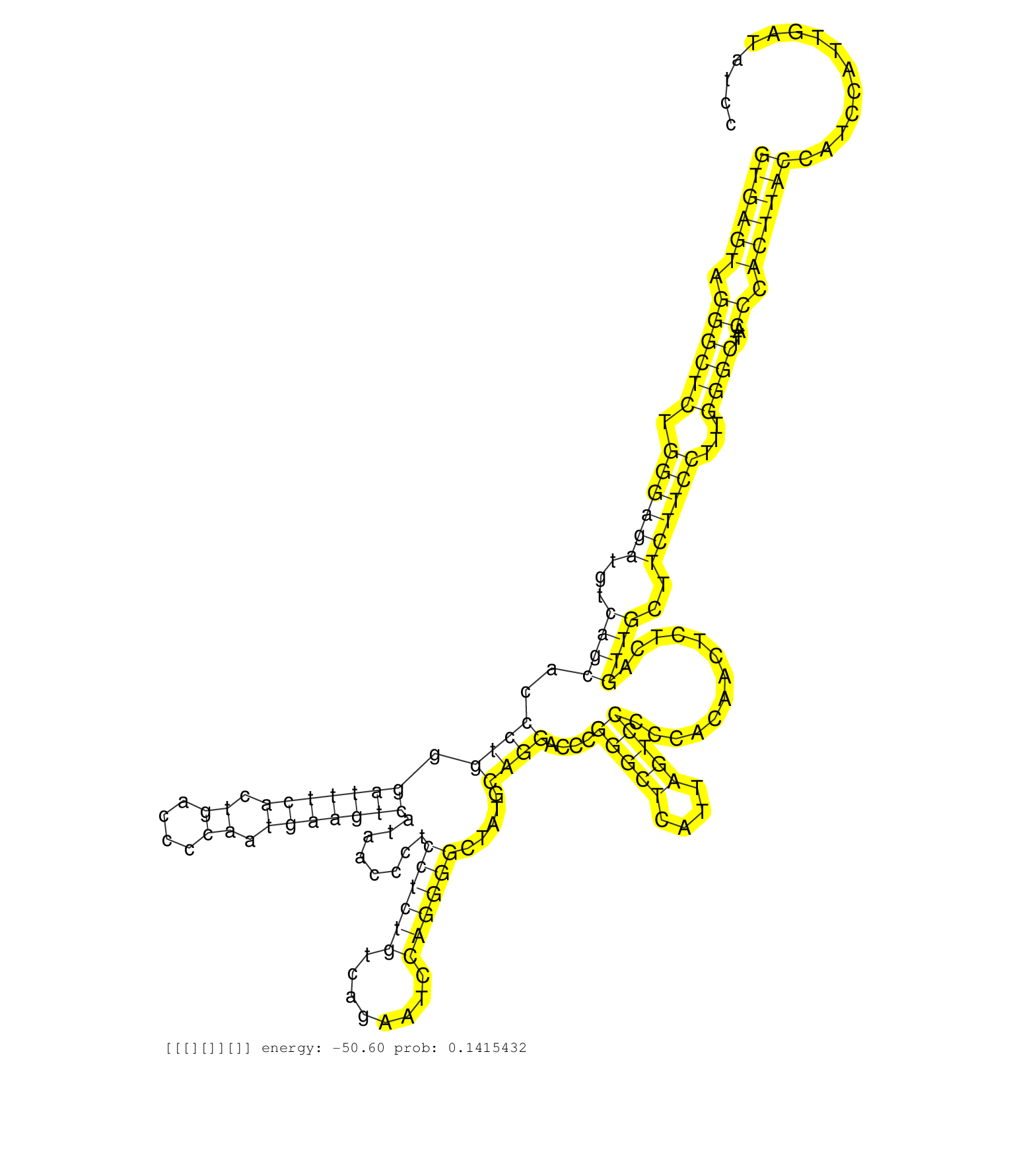

| Gene: Cyp11a1 | ID: uc009pwa.1_intron_7_0_chr9_57874090_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) PIWI.ip |
(14) TESTES |
| GTCTGGGCCGGCGGATTGCGGAGCTGGAGATGACCATCCTCCTTATCAATGTGAGTAGGGCTCTGGGAGATGTCAGCACCCTGGGATTTCACTGACCCCAATGAAGTCATAACCCTCCTCTGTCAGAATCCAGGGGCTATGCAGGACCCGGGCTCATTAGTCCCCCACAACTCTCAGTTGCTTCTTCCTTTGGGCTTACCCACTTACCATCCATTGATATCCCCTAAACACTTATCTGCCCCCACTTGGGAGGTGATGTGGGAGAGATTGCTACAGACTAGTTAAAGTCAAAGGCTGAATCCATGAAACAGAGCATGCAGGGACCATTTAGACCACAGTCAAGAGGTTGAATAGTTTGTGTCCTTTGGATCACACTCAAGAGATTGACCCTGTAGCTACAGATGTCATGGGTGTGTGACCCAGGAGAGACAAATCCTCCTTTCTGCCTCCTTCTTCTTCAGCTGCTGGAGAACTTCAGAATTGAAGTTCAAAATCTCCGTGATGTGGGGAC ..................................................((((((.((((((.((((((...((((..((((.(((((((.((....)).)))))))........((((((........)))))).....))))....(((((....))))).............))))..))))))...))))...)).))))))................................................................................................................................................................................................................................................................................................................ ..................................................51.........................................................................................................................................................................222............................................................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................AATCCAGGGGCTATGCAGGACCCGGGCTCATTAGTCCCCCACAACTCTCAGT............................................................................................................................................................................................................................................................................................................................................. | 52 | 1 | 89.00 | 89.00 | 89.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....GGGCCGGCGGATTGCGGAGCTG..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTGCGGAGCTGGAGATct.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | ct | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - |
| ...........CGGATTGCGGAGCTGGAG.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CGGATTGCGGAGCTGGAGATGA.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................TGCGGAGCTGGAGATGACCATCCTCCT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........GCGGATTGCGGAGCTGGAGA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .TCTGGGCCGGCGGATTGCGGA......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGCGGAGCTGGAGATGACC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGAGAACTTCAGAATTGA........................... | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........CGGATTGCGGAGCTGGAGATGACC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .TCTGGGCCGGCGGATTGCGGAGCTGGAGATGAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGCTGGAGAACTTCAGAATTGAAG......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................TGCGGAGCTGGAGATGACCA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGTAGGGCTCcacc............................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | cacc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........CGGATTGCGGAGCTGGAGATG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................GCGGAGCTGGAGATGACt............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........GCGGATTGCGGAGCTGGAGATGACCAT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAAATCTCCGTGATGTGGG... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............GATTGCGGAGCTGGAGATGACCta.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | ta | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................TGCGGAGCTGGAGATGACCATCCTaga.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................ACCCTCCTCTGTCAGAgaa............................................................................................................................................................................................................................................................................................................................................................................................. | 19 | gaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............TTGCGGAGCTGGAGATGAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTGAAGTTCAAAATCTCCGTGATGTGG.... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................GCGGAGCTGGAGATGACCA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGTAGGGCTCaaac............................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | aaac | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................TCCTTCTTCTTCAGCTGgta............................................ | 20 | gta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....GGGCCGGCGGATTGCGGAGCTGGAGATGACCAT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTGCTGGAGAACTTCAGA................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .TCTGGGCCGGCGGATTGC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........GCGGATTGCGGAGCTGGAGATGAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........GCGGATTGCGGAGCTGGAGATGACC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......CCGGCGGATTGCGGAGCTGGAGATGACCAT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................TGAAACAGAGCATGCAGG.............................................................................................................................................................................................. | 18 | 4 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ...........CGGATTGCGGAGCTG..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTTCTTCAGCTGCTGGA.......................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| GTCTGGGCCGGCGGATTGCGGAGCTGGAGATGACCATCCTCCTTATCAATGTGAGTAGGGCTCTGGGAGATGTCAGCACCCTGGGATTTCACTGACCCCAATGAAGTCATAACCCTCCTCTGTCAGAATCCAGGGGCTATGCAGGACCCGGGCTCATTAGTCCCCCACAACTCTCAGTTGCTTCTTCCTTTGGGCTTACCCACTTACCATCCATTGATATCCCCTAAACACTTATCTGCCCCCACTTGGGAGGTGATGTGGGAGAGATTGCTACAGACTAGTTAAAGTCAAAGGCTGAATCCATGAAACAGAGCATGCAGGGACCATTTAGACCACAGTCAAGAGGTTGAATAGTTTGTGTCCTTTGGATCACACTCAAGAGATTGACCCTGTAGCTACAGATGTCATGGGTGTGTGACCCAGGAGAGACAAATCCTCCTTTCTGCCTCCTTCTTCTTCAGCTGCTGGAGAACTTCAGAATTGAAGTTCAAAATCTCCGTGATGTGGGGAC ..................................................((((((.((((((.((((((...((((..((((.(((((((.((....)).)))))))........((((((........)))))).....))))....(((((....))))).............))))..))))))...))))...)).))))))................................................................................................................................................................................................................................................................................................................ ..................................................51.........................................................................................................................................................................222............................................................................................................................................................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................................................................................................................................CACACTCAAGAGATTGtgt............................................................................................................................. | 19 | tgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................CTGAATCCATGAAACata.......................................................................................................................................................................................................... | 18 | ata | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................CTCTGGGAGATGTCAttgg.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | ttgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |