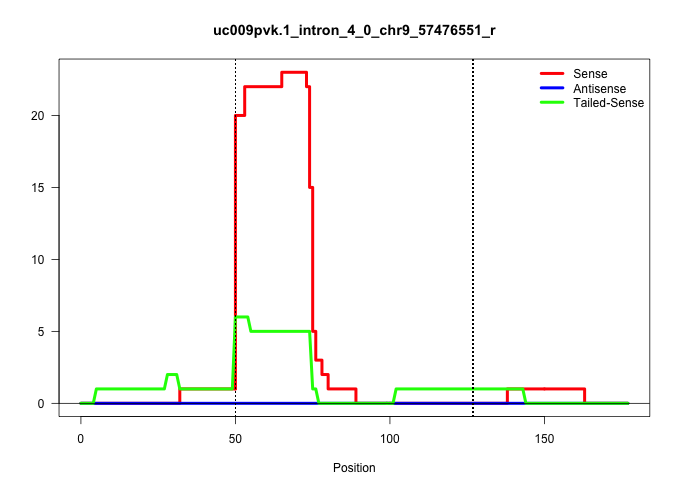
| Gene: Csk | ID: uc009pvk.1_intron_4_0_chr9_57476551_r | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| AAGAATGACGCAACTGCCCAGGCCTTCCTGGCTGAAGCCTCCGTCATGACGTGAGCTGGACTGCCTGGGTGGAGCACCGGGTGGGGGGTTGGCAGGCCGAACTCCTGACCACCCTGTCCTGCCACAGGCAACTTCGGCACAGCAACCTCGTCCAGCTGCTGGGTGTGATTGTGGAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGC...................................................................................................... | 25 | 1 | 10.00 | 10.00 | 3.00 | - | 2.00 | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAG....................................................................................................... | 24 | 1 | 7.00 | 7.00 | - | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGg...................................................................................................... | 25 | g | 2.00 | 7.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGCA..................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGCACC................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TCCTGACCACCCTGTCCTGCCACAGt................................................. | 26 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CAACTTCGGCACAcct................................. | 16 | cct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGaag.................................................................................................... | 27 | aag | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGt...................................................................................................... | 25 | t | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGCTGGACTGCCTGGGTGGAGa...................................................................................................... | 25 | a | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TGACGCAACTGCCCAGGCCTTCCTGtt................................................................................................................................................. | 27 | tt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACAGCAACCTCGTCCAGCTGCTGGG.............. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................TGGGTGGAGCACCGGGTGGGGGGT........................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGCTGGACTGCCTGGGTGGAGCACCGG................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................AGCTGGACTGCCTGGGTGGA........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAAGCCTCCGTCATGAC............................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................TGGCTGAAGCCTCCGTCATGACGcaac.......................................................................................................................... | 27 | caac | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| AAGAATGACGCAACTGCCCAGGCCTTCCTGGCTGAAGCCTCCGTCATGACGTGAGCTGGACTGCCTGGGTGGAGCACCGGGTGGGGGGTTGGCAGGCCGAACTCCTGACCACCCTGTCCTGCCACAGGCAACTTCGGCACAGCAACCTCGTCCAGCTGCTGGGTGTGATTGTGGAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................GGGTTGGCAGGCCGtgga.............................................................................. | 18 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................CCTCGTCCAGCTGCcgc.................. | 17 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................GGGTTGGCAGGCCGAtgga............................................................................. | 19 | tgga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |