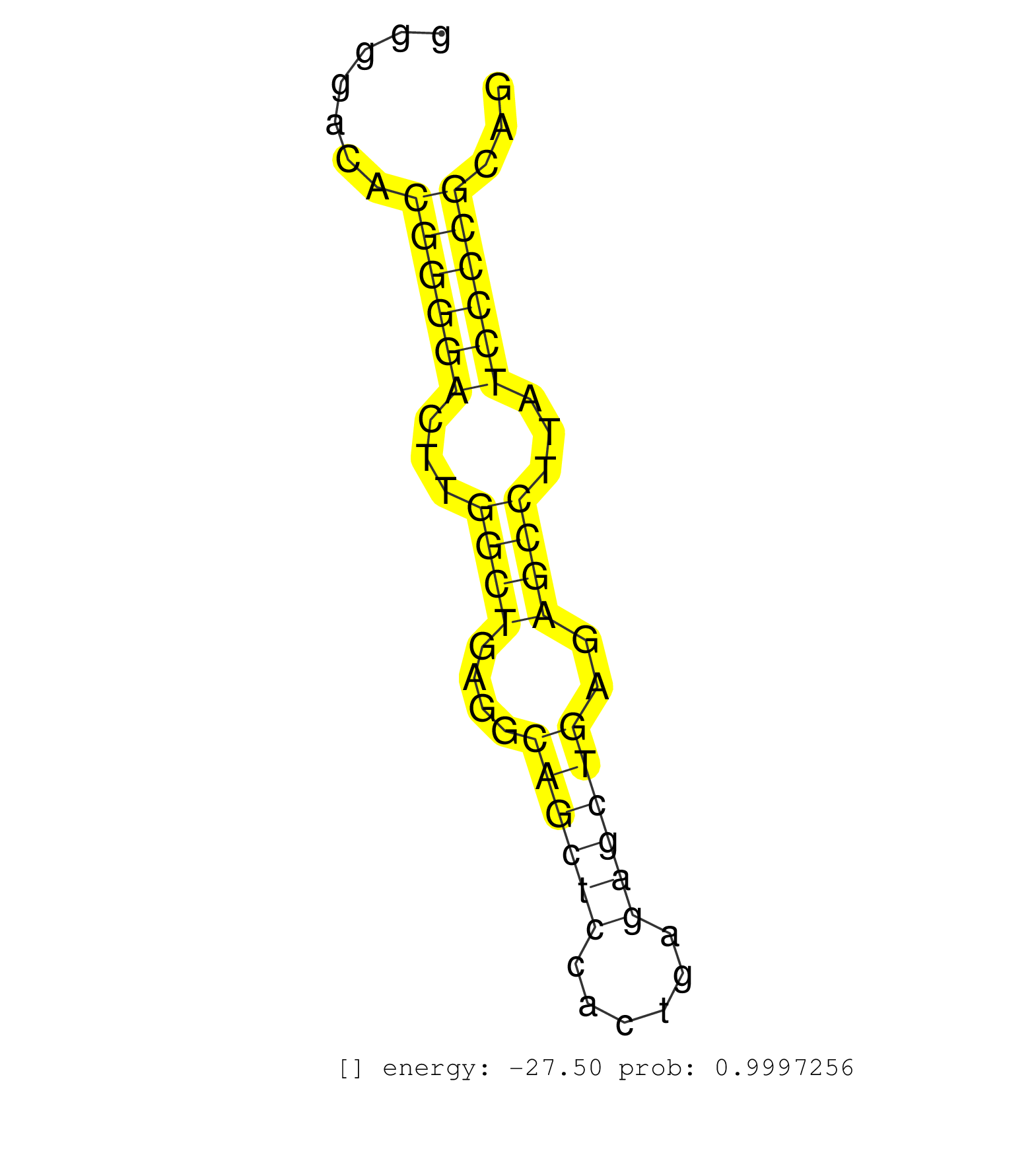
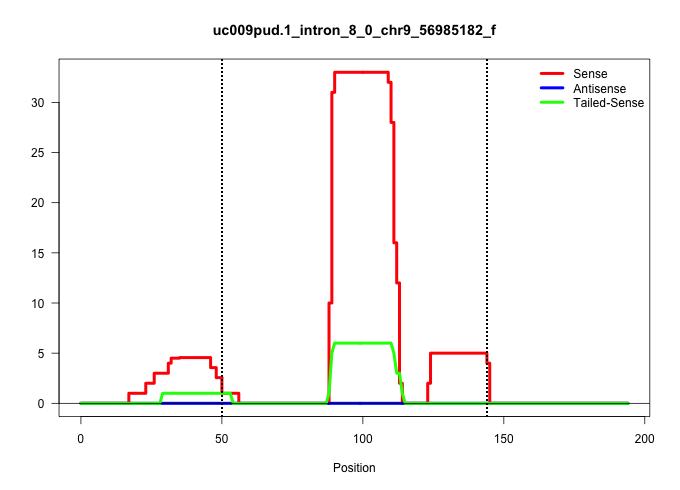
| Gene: Man2c1 | ID: uc009pud.1_intron_8_0_chr9_56985182_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(23) TESTES |
| TGCAGGGCCAGAACTTCTTTCTGCAAGAGTTTGGGAAGATGTGCTCTGAGGTAGTCTGGGAGCTGGCACCGCAGACTATCCCCAGGGGACACGGGGACTTGGCTGAGGCAGCTCCACTGAGAGCTGAGAGCCTTATCCCCGCAGTTCTGGCTGCCAGACACGTTTGGCTACTCAGCACAGCTCCCCCAGATCAT ...........................................................................................((((((...((((....((((((......))))))..))))...))))))..................................................... ....................................................................................85.........................................................144................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................CACGGGGACTTGGCTGAGGCAGCT................................................................................. | 24 | 1 | 50.00 | 50.00 | 39.00 | - | - | 2.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCA.................................................................................... | 21 | 1 | 33.00 | 33.00 | 22.00 | - | - | - | 7.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCAGC.................................................................................. | 24 | 1 | 28.00 | 28.00 | 13.00 | - | 3.00 | 6.00 | - | - | 1.00 | 1.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAG................................................................................... | 22 | 1 | 23.00 | 23.00 | - | 10.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCAG................................................................................... | 23 | 1 | 10.00 | 10.00 | - | 4.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGa.................................................................................. | 23 | a | 7.00 | 23.00 | - | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCA.................................................................................... | 22 | 1 | 6.00 | 6.00 | - | - | 2.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGt.................................................................................. | 23 | t | 6.00 | 23.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAGAGCCTTATCCCCGCAGT................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTGAGAGCCTTATCCCCGCAGT................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| ............................................................................................GGGGACTTGGCTGAGGCAGCTC................................................................................ | 22 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCAt................................................................................... | 23 | t | 2.00 | 6.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GGGAAGATGTGCTCTGAG................................................................................................................................................ | 18 | 2 | 2.00 | 2.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGCTC................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAt................................................................................... | 22 | t | 2.00 | 33.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GACACGGGGACTTGGCTGAGGCAGC.................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCAGCT................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................GAGTTTGGGAAGATGTGCTCTG.................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGC..................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGC.................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCAat.................................................................................. | 24 | at | 1.00 | 6.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTGAGAGCCTTATCCCCGCAGTat............................................... | 24 | at | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................ACGGGGACTTGGCTGAGGCAGCTC................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGCTatat............................................................................. | 28 | atat | 1.00 | 50.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TTTGGGAAGATGTGCTCTGAGttct............................................................................................................................................ | 25 | ttct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGCTa................................................................................ | 25 | a | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................ACGGGGACTTGGCTGAGGCAGCT................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGGGAAGATGTGCTCTGAGGTAGTC.......................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................CAAGAGTTTGGGAAGATGTGCTCTGAG................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGga.................................................................................... | 21 | ga | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAGAGTTTGGGAAGATGTGCTCTGAG................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................TTTCTGCAAGAGTTTGGGAAGATGTGCTC.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................ACGGGGACTTGGCTGAGGCAGaaga............................................................................... | 25 | aaga | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CACGGGGACTTGGCTGAGGCAGCTt................................................................................ | 25 | t | 1.00 | 50.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGCt.................................................................................... | 22 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTGAGAGCCTTATCCCCGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................ACACGGGGACTTGGCTGAGGC..................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................ACGGGGACTTGGCTGAGGCAG................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................AAGATGTGCTCTGAG................................................................................................................................................ | 15 | 17 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - |
| TGCAGGGCCAGAACTTCTTTCTGCAAGAGTTTGGGAAGATGTGCTCTGAGGTAGTCTGGGAGCTGGCACCGCAGACTATCCCCAGGGGACACGGGGACTTGGCTGAGGCAGCTCCACTGAGAGCTGAGAGCCTTATCCCCGCAGTTCTGGCTGCCAGACACGTTTGGCTACTCAGCACAGCTCCCCCAGATCAT ...........................................................................................((((((...((((....((((((......))))))..))))...))))))..................................................... ....................................................................................85.........................................................144................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|