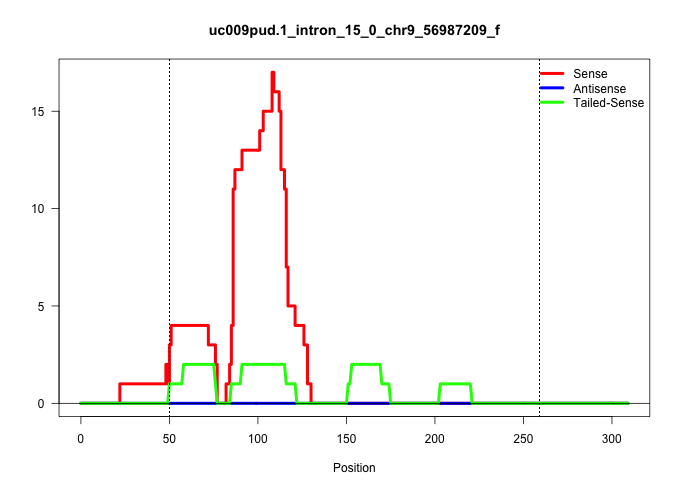
| Gene: Man2c1 | ID: uc009pud.1_intron_15_0_chr9_56987209_f | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(4) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
| GCGCACTGAAGTGTTGGCACTGCCCAAGCCTTGTGGGGCTCACAGCCTAGGTATGAGCTGACAGGGGGAAGCTGGTCTTGCAGCAGGAAAATCGAAGGCTAGACAGCATGACAAGGATACTCCCCAGGGAATCTTTACCTTGGGGTATCCCAATCTGGAGAGAAGTAGCCGTGTACTATCCCATCCAGCCTATCCTTAACCTGAGCAAGGTGTCTAGACCTTCAGAGAGGCTACACTGATGCCTAGGCCTTGTCCCCAGCCCTGGTGACAGTGCCAAGTATTGGCTACGCTCCTGCTCCCACCCCTACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................GAAAATCGAAGGCTAGACAGCATGACAAGG................................................................................................................................................................................................. | 30 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAAAATCGAAGGCTAGACAGCATGACA.................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TGACAAGGATACTCCCCAGG..................................................................................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAAAATCGAAGGCTAGACAGCATGACAAGGA................................................................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCGAAGGCTAGACAGCATGACAAGGATACTt........................................................................................................................................................................................... | 31 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GGAAAATCGAAGGCTAGACAGCATGACAAGt................................................................................................................................................................................................. | 31 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGAGCTGACAGGGGGAAGCTGGTC........................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GCAGGAAAATCGAAGGCTAGACAGCAT........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................AGGAAAATCGAAGGCTAGACAGCATGACA.................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TATGAGCTGACAGGGGGAAGCTGGTC........................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TCGAAGGCTAGACAGCATGACAAGGATACT............................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGACAGGGGGAAGCacga......................................................................................................................................................................................................................................... | 18 | acga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................GACAGCATGACAAGGATACTCCCCA....................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................CCCAAGCCTTGTGGGGCTCACAGCCTA.................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................GGAAAATCGAAGGCTAGACAGCATGACAAG.................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................AAAATCGAAGGCTAGACAGCATGACAAGGA................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGCATGACAAGGATACTCCCCAGGGA................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AGCAAGGTGTCTAGcatg........................................................................................ | 18 | catg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTATGAGCTGACAGGGGGAAGC............................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................GGAAAATCGAAGGCTAGACAGCATGACAAGG................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................AATCTGGAGAGAAGTcatg........................................................................................................................................... | 19 | catg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TCTGGAGAGAAGTAGCCGcgta...................................................................................................................................... | 22 | cgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTATGAGCTGACAGGGGGAAGCTGGTa........................................................................................................................................................................................................................................ | 27 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTATGAGCTGACAGGGGGAAGCTGGT......................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GAAAATCGAAGGCTAGACAGCATGAC..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| GCGCACTGAAGTGTTGGCACTGCCCAAGCCTTGTGGGGCTCACAGCCTAGGTATGAGCTGACAGGGGGAAGCTGGTCTTGCAGCAGGAAAATCGAAGGCTAGACAGCATGACAAGGATACTCCCCAGGGAATCTTTACCTTGGGGTATCCCAATCTGGAGAGAAGTAGCCGTGTACTATCCCATCCAGCCTATCCTTAACCTGAGCAAGGTGTCTAGACCTTCAGAGAGGCTACACTGATGCCTAGGCCTTGTCCCCAGCCCTGGTGACAGTGCCAAGTATTGGCTACGCTCCTGCTCCCACCCCTACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................GGATACTCCCCAccgc....................................................................................................................................................................................... | 16 | ccgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................CCAGCCTATCCTTAggg............................................................................................................... | 17 | ggg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |