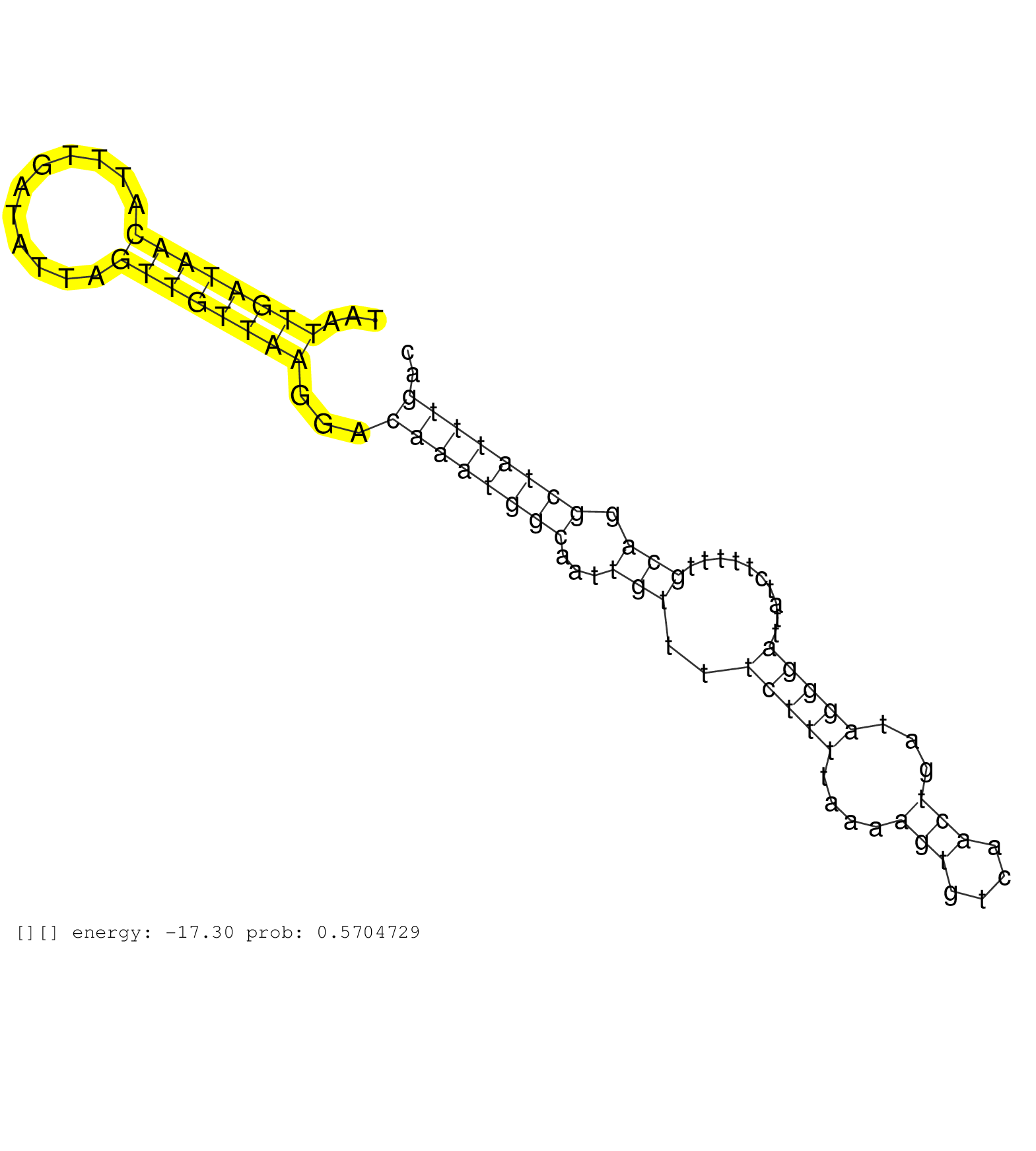
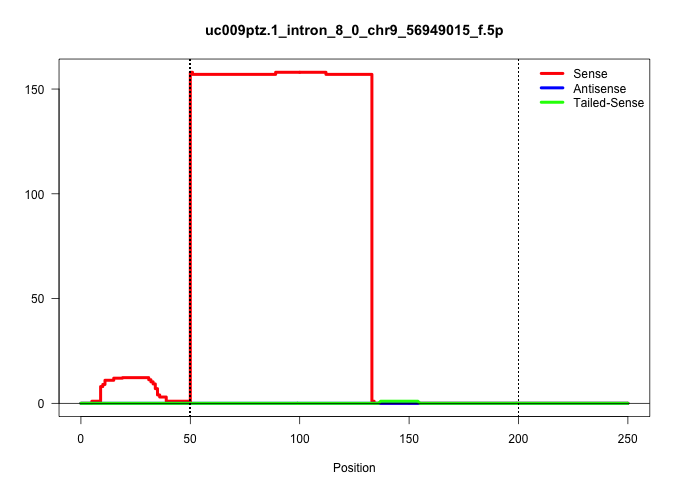
| Gene: Sin3a | ID: uc009ptz.1_intron_8_0_chr9_56949015_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| CAGATGCCAGCAAGCATGGTGTTGGAACGGAATCATTATTTTTTGATAAGGTAAGTAATTGGTGACATTTGATATTTTTGTTAAAGACAAATGGCATAAGTAATTGATAACATTTGATATTAGTTGTTAAGGACAAATGGCAATTGTTTTCTTTTAAAAGTGTCAACTGATAGGGATTATCTTTTTGCAGGCTATTTGACTTTTGTTTAATTTGGAGACAGACTCTATATAACCCAGGTTGCCTTTGAAC .......................................................................................................((((((((...........))))))))...((((((((...(((..(((((....(((....)))...)))))..........))).)))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTAATTGGTGACATTTGATATTTTTGTTAAAGACAAATGGCATAAGTA.................................................................................................................................................... | 52 | 1 | 163.00 | 163.00 | 163.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GCAAGCATGGTGTTGGAACGGAATC........................................................................................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........GCAAGCATGGTGTTGGAACGGAATCA....................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| .................................CATTATTTTTTGATAAGG....................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............ATGGTGTTGGAACGGAAT......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....GCCAGCAAGCATGGTGTTGGAACGGA........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................AATGGCATAAGTAATTGATAACA.......................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........AAGCATGGTGTTGGAACGGAATCA....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........CAAGCATGGTGTTGGAACGGAA.......................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGGCAATTGTTTTCatct............................................................................................... | 18 | atct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........AAGCATGGTGTTGGAACGGAATCATTAT................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........GCAAGCATGGTGTTGGAACGGAATCATTAT................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........GCAAGCATGGTGTTGGAACGGAAT......................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........GCAAGCATGGTGTTGGAACGGAATCAT...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................TGTTGGAACGGAATC........................................................................................................................................................................................................................ | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| CAGATGCCAGCAAGCATGGTGTTGGAACGGAATCATTATTTTTTGATAAGGTAAGTAATTGGTGACATTTGATATTTTTGTTAAAGACAAATGGCATAAGTAATTGATAACATTTGATATTAGTTGTTAAGGACAAATGGCAATTGTTTTCTTTTAAAAGTGTCAACTGATAGGGATTATCTTTTTGCAGGCTATTTGACTTTTGTTTAATTTGGAGACAGACTCTATATAACCCAGGTTGCCTTTGAAC .......................................................................................................((((((((...........))))))))...((((((((...(((..(((((....(((....)))...)))))..........))).)))))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................TTTTGTTAAAGACAccct................................................................................................................................................................. | 18 | ccct | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TCTTTTAAAAGTGTCttta...................................................................................... | 19 | ttta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |